Cellpose Segmentation¶
This tool uses 2.0 compute credits per hour.
Overview¶
The Cellpose Segmentation tool provides cell segmentation using Cellpose with advanced features tailored for the IDEAS Toolbox. It supports image preprocessing (LoG filtering), handles multiple file formats including ISXD, and offers flexible output options such as NPY, ISXD, and TIFF stacks. The tool also generates comprehensive previews with ROI overlays and statistics. For more information on the underlying algorithm, visit the Cellpose website.
Input Data¶
This tool accepts various image formats for segmentation.
Valid Inputs¶
| File Type | Required? | Description |
|---|---|---|
| Image Files | Yes | Primary workflow expects a single image file per run. Supported formats include TIFF (.tif, .tiff), PNG (.png), ISXD (.isxd), and NPY (.npy). |
Parameters¶
| Parameter | Required? | Default | Description |
|---|---|---|---|
| Input Image Files | True | N/A | List of input image file paths for segmentation. Supports multiple formats including TIFF stacks, PNG, NumPy arrays, and ISXD files (INSCOPIX calcium imaging data). Images should contain cells or cell-like structures for optimal segmentation results. |
| Model Type | False | cpsam | Cellpose model architecture to use for segmentation. 'cpsam' (CellposeSAM) provides state-of-the-art performance with automatic diameter estimation. 'custom' allows use of user-trained models (requires custom_model_path). |
| Expected Cell Diameter | True | N/A | Expected cell diameter in pixels. Leave blank for automatic detection (recommended for cpsam model). For manual specification: measure typical cell diameter in your images. Smaller values detect smaller objects, larger values detect bigger objects. |
| Flow Threshold | True | 1.0 | Flow error threshold for mask creation. Lower values (0.1-0.3) create more masks, higher values (0.5-1.0) create fewer, more confident masks. Default 0.4 works well for most cell types. |
| Cell Probability Threshold | True | -1.6 | Cell probability threshold for mask creation. Range: -6.0 to 6.0. Negative values are more sensitive (detect more cells), positive values are more conservative. -1.0 to -2.0 good for detecting faint cells, 0.0 to 1.0 for clear cells only. |
| Minimum Cell Size | False | 30 | Minimum size of segmented objects in pixels. Objects smaller than this will be excluded from results. Set based on smallest real cells in your images to filter out noise and debris. |
| Preprocessing Cell Diameter | False | 0 | Expected cell diameter in pixels for LoG convolution preprocessing. Set to 0 to disable preprocessing. When >0, enables Laplacian of Gaussian filtering using this diameter for enhanced cell detection. Should match typical cell size in your images. |
| Apply Tiling | False | True | Apply tiling by dividing input images into overlapping tiles. When enabled, the function divides images into overlapping tiles (default 64×64 pixels), predicts labels for each tile individually, and averages results for overlapping regions. This matches MATLAB's Tile parameter behavior and is recommended for large images. |
| Tile Overlap | False | 0.1 | Overlap fraction for tiling. Controls the amount of overlap between tiles. For most images, suitable values range from 0.1 (10% overlap) to 0.5 (50% overlap). Higher values improve edge accuracy but increase computation time. This matches MATLAB's TileOverlap parameter. |
| 3D Processing | False | False | Enable 3D segmentation for image stacks. Use for Z-stack or time-series data where cells span multiple planes. When enabled, the model processes the entire volume considering inter-slice relationships. |
| Slice Count Control for 3D Processing | False | N/A | Slice range for 3D processing. Must be two integers [start, end] for specific slice range (e.g., [10, 50] to process slices 10 through 50, inclusive), or blank/null to process all slices. Indices are 0-based (first slice is 0). Examples: [0, 9] processes first 10 slices, [20, 29] processes slices 20-29. This helps manage memory usage and processing time for large z-stacks. |
| Z-Axis | False | 0 | Which axis represents the z-dimension for 3D processing. 0 = first axis (typical for z-stacks), 1 = second axis, 2 = third axis. Only used when 3D Processing is enabled. |
| Channel Axis | False | -1 | Which axis represents the color channels for 4D processing. -1 = last axis (typical for RGB), 0 = first axis, 1 = second axis, etc. Only used when 3D Processing is enabled for 4D images. |
| Save Masks | False | N/A | Enable saving of segmentation masks to output files. When enabled, masks will be saved in the format specified by output_format parameter. |
| Save Statistics | False | N/A | Enable saving of cell statistics to CSV files. When enabled, individual cell morphology and intensity statistics will be generated. |
| Output Format | True | npy | Select output format for detected ROI/mask data only. NPY provides fastest I/O and compatibility, ISXD enables INSCOPIX calcium imaging workflows, TIFF stacks offer broad compatibility with image analysis software. Note: Statistics (CSV), metadata (JSON), and preview images (PNG) are always generated in their standard formats regardless of this setting. |
| GPU Processing Mode | False | True | Enable GPU processing for accelerated segmentation. When enabled, uses GPU if available (CUDA/MPS), otherwise falls back to CPU. GPU processing significantly accelerates segmentation for large images. |
| Tile Size | False | 256 | Size of image tiles in pixels for processing (when tile=True). Default changed to 64×64 for maximum compatibility across different image sizes and hardware configurations. Smaller values reduce memory usage and improve compatibility, while larger values may be faster but can cause tensor dimension errors on some systems. |
| Batch Size | False | 1 | Number of images to process simultaneously. Higher values use more memory but may be faster for multiple images. |
Algorithm Details¶
The tool follows a structured workflow for segmentation:
Key Features¶
- Preprocessing: Optional Laplacian of Gaussian (LoG) filtering to enhance cell boundaries before segmentation.
- Multi-Format Support:
- Input: Handles standard image formats and specialized ISXD files.
- Output: Choose one target mask format per run (
output_formatacceptsnpy,isxd, ortiff_stackfor 2D data; 3D data must usenpy;isxdoutput requiresisxdinput).
- 3D Processing: Specialized handling for z-stacks, including 4D output format (HxWxcellxslice) for 3D consistency.
- CPSAM Model Optimization: Optimized for the
cpsammodel with automatic GPU detection.
3D Processing (Z-Stacks)¶
The tool supports processing of 3D volumetric data (z-stacks).
- Activation: Set
do_3Dtotrue. The tool will automatically process the data as a volume. - Axis Configuration:
z_axis: Specifies the dimension index corresponding to the z-stack (depth). Default is0.channel_axis: Specifies the dimension index for color channels. Default is-1(last dimension).- Note: Incorrect axis settings can lead to errors or incorrect segmentation (e.g., processing slices as channels).
- Slicing: You can limit processing to a specific range of slices using
max_slices_3d.- Format:
[start_index, end_index](e.g.,[10, 50]). Indices are 0-based and inclusive.
- Format:
- Potential Errors:
- Output Format: 3D processing produces 4D output (HxWxcellxslice) which cannot be saved as a standard TIFF file. You MUST set
output_formattonpywhendo_3Dis enabled. - Dimensionality Mismatch: If
do_3Distruebut the input image is 2D, the tool will raise an error. Conversely, if the input is clearly a z-stack butdo_3Disfalse, the tool may error or produce unexpected results depending on the exact dimensions.
- Output Format: 3D processing produces 4D output (HxWxcellxslice) which cannot be saved as a standard TIFF file. You MUST set
Parameter Optimization Tips¶
- Diameter: Start with the default (30) and adjust based on your cell size. Setting to 0 enables automatic diameter estimation (slower but potentially more accurate).
- Thresholds:
- Flow Threshold: Default is 1.0. Increase (e.g., 2.0) to be more permissive (more ROIs); decrease (e.g., 0.4) to be more stringent.
- Cell Probability Threshold: Default is -1.6. Decrease (e.g., -2.0) to be more permissive (find fainter cells); increase (e.g., 0.0) to be more stringent.
- Preprocessing: Enable for noisy or low-contrast images.
- Large Images: Use tiling (
tile=true) for images larger than 2000x2000 pixels to manage memory usage. - Performance: Enable GPU (
use_gpu=true) for faster processing when available. - Block Size (
bsize): Ensurebsizeis compatible with your image dimensions (frame size).- Critical for CPSAM: The app may fail with an error if
bsizedoes not match or is incompatible with the frame size. - Recommendation: If you encounter runtime errors, try setting
bsizeto match your image dimension (e.g., 512 for 512x512 images) or use standard powers of 2 (256, 512).
- Critical for CPSAM: The app may fail with an error if
Example Configurations¶
High-quality cells, good contrast:
{
"diameter": 25,
"flow_threshold": 0.4,
"cellprob_threshold": 0.0,
"preprocessing_cell_diameter": 0
}
Noisy images, poor contrast:
{
"diameter": 30,
"flow_threshold": 0.8,
"cellprob_threshold": -2.0,
"preprocessing_cell_diameter": 25
}
Large images, memory constraints:
{
"tile": true,
"tile_overlap": 0.2,
"bsize": 128,
"use_gpu": true
}
Outputs¶
The tool generates segmentation masks, statistics, and visualization previews.
Data Files¶
individual_statistics.csv(primary output): Morphological and intensity statistics for each detected cell whensave_statistics=true.cellpose_masks: Segmentation masks saved in the format specified byoutput_formatwhensave_masks=true. ISXD export (.isxd) requires an ISXD input file. 3D datasets are exported only as.npy.
Previews¶
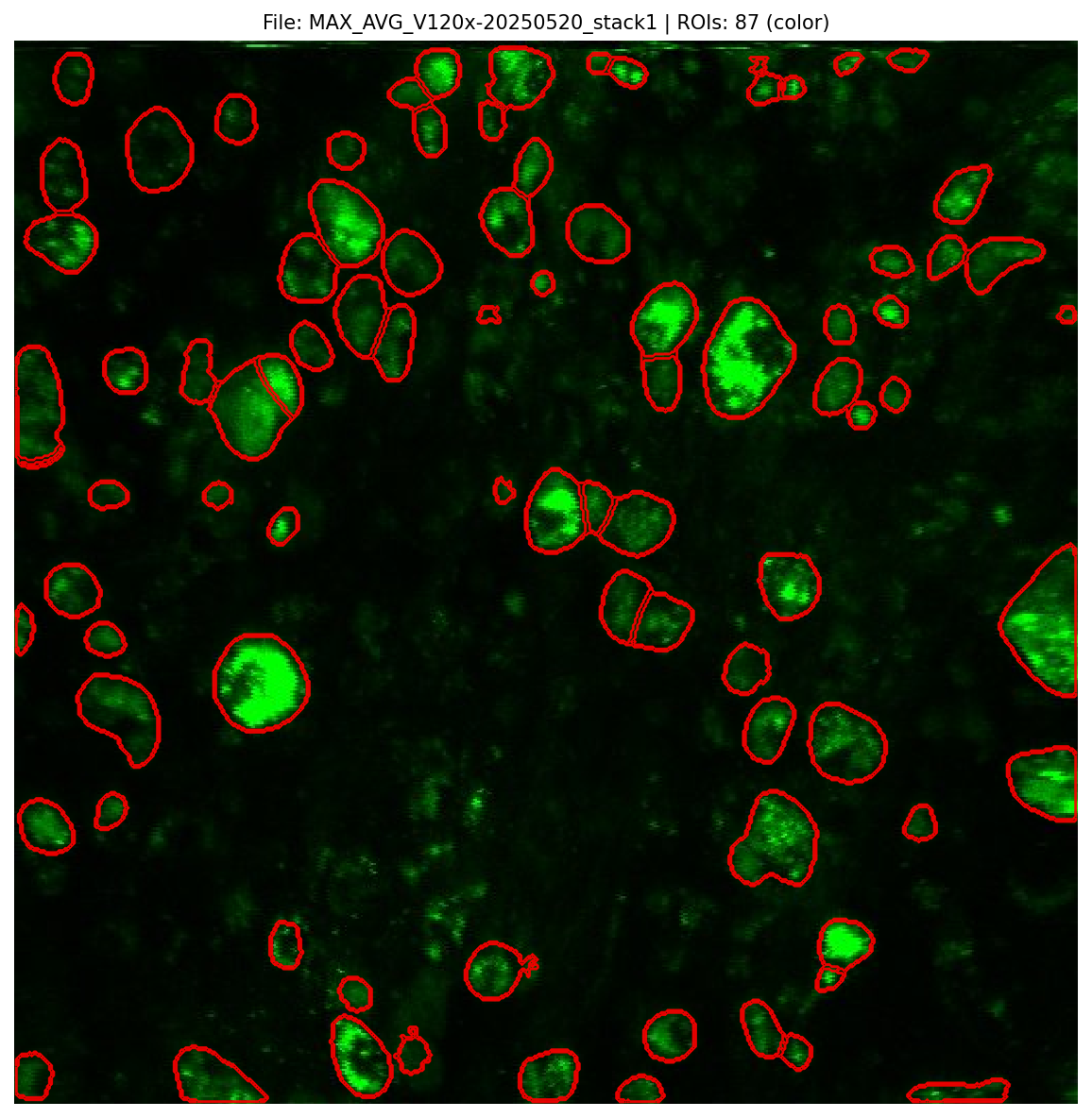
Segmentation Preview¶
The image below shows an example preview with the original image, ROI overlays, and summary statistics.