suite2p ROI extraction¶
This tool uses 1.0 compute credits per hour.
Overview¶
This suite2p ROI extraction tool is a wrapper for the suite2p function extraction.extraction_wrapper().
You can find more information on this processing step in the suite2p docs.
suite2p ROI extraction extracts neuropil-corrected fluorescence traces and corresponding neuropil fluorescence traces from the registered movie for all detected ROIs.
Note that this tool constitutes the fourth step of Run suite2p pipeline.
Input files¶
| Source Parameter | File Type | File Format |
|---|---|---|
| Registered Binary Movie | suite2p_data | bin |
| Detection Statistics File | suite2p_data | npy |
| Parameters File | config | npy |
Parameters¶
| Parameter | suite2p name | Required? | Default | Description |
|---|---|---|---|---|
| Registered Binary Movie | - | True | N/A | Input registered suite2p binary movie [.bin] |
| Detection Statistics File | - | True | N/A | Input cell statistics file, as outputted by the ROI detection step [ |
| Parameters File | - | True | N/A | Input suite2p parameters file, as outputted by the ROI detection tool [ |
| Neuropil Extraction | neuropil_extract | False | True | [from suite2p docs] "Whether or not to extract signal from neuropil. If False, Fneu is set to zero." [bool] |
| Allow Overlap | allow_overlap | False | False | [from suite2p docs] "Whether or not to extract signals from pixels which belong to two ROIs. By default, any pixels which belong to two ROIs (overlapping pixels) are excluded from the computation of the ROI trace." [bool] |
| Minimum Neuropil Pixels | min_neuropil_pixels | False | 350 | [from suite2p docs] "Minimum number of pixels used to compute neuropil for each cell." [int, 0] |
| Inner Neuropil Radius | inner_neuropil_radius | False | 2 | [from suite2p docs] "Number of pixels to keep between ROI and neuropil donut." [int, =0] |
| Lambda Percentile | lam_percentile | False | 50 | [from suite2p docs] "Percentile of Lambda within area to ignore when excluding cell pixels for neuropil extraction." [int, [0 100]] |
| Show Grid on FOV | - | False | True | Whether or not to show the grid on FOVs [bool] |
| FOV Ticks Step | - | False | 128 | Step for the x- and y-ticks [int, >0] |
| Number of Sample Cells | - | False | 20 | Number of sample cells for the cell extraction preview [int, >0] |
| Random Seed | - | False | 0 | Random seed for selecting sample cells [int, >=0] |
| Show Non-Sample Cells' Footprints | - | False | True | Whether or not to show footprints of non-sample cells on the cell footprint FOV image. If False, only footprints of sample cells are displayed [bool] |
Output files¶
| File name | File type | Notes |
|---|---|---|
| stat.npy | NumPy file | Contains spatial and functional statistics for all extracted ROIs, as a dictionary. |
| F.npy | NumPy file | Contains neuropil-corrected fluorescence traces for all extracted ROIs, as a 2D array. |
| Fneu.npy | NumPy file | Contains neuropil fluorescence traces for all extracted ROIs, as a 2D array. |
| ops_ROI_extraction.npy | NumPy file | Contains a dictionary of parameters populated by suite2p ROI extraction. |
You can explore 1 preview figures for stat.npy and 2 for F.npy.
You can find below an example of these previews obtained from processing a 5-minute 2P movie of mouse cortex (data courtesy of Dr. Ahmet Arac, MD, at UCLA).
For stat.npy:
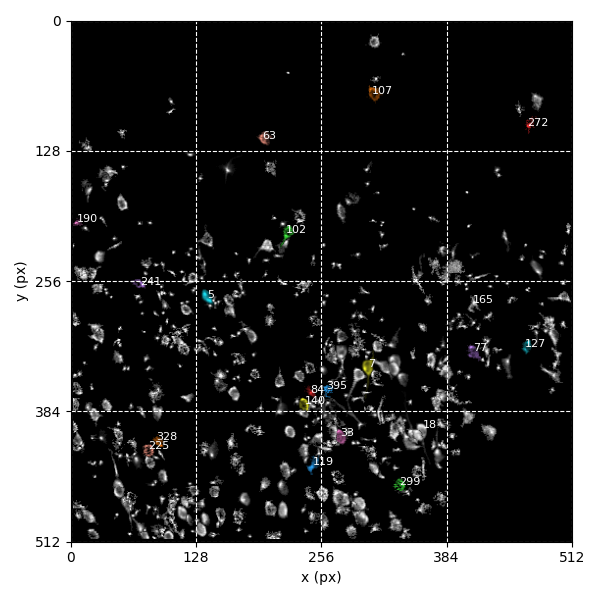 |
and for F.npy:
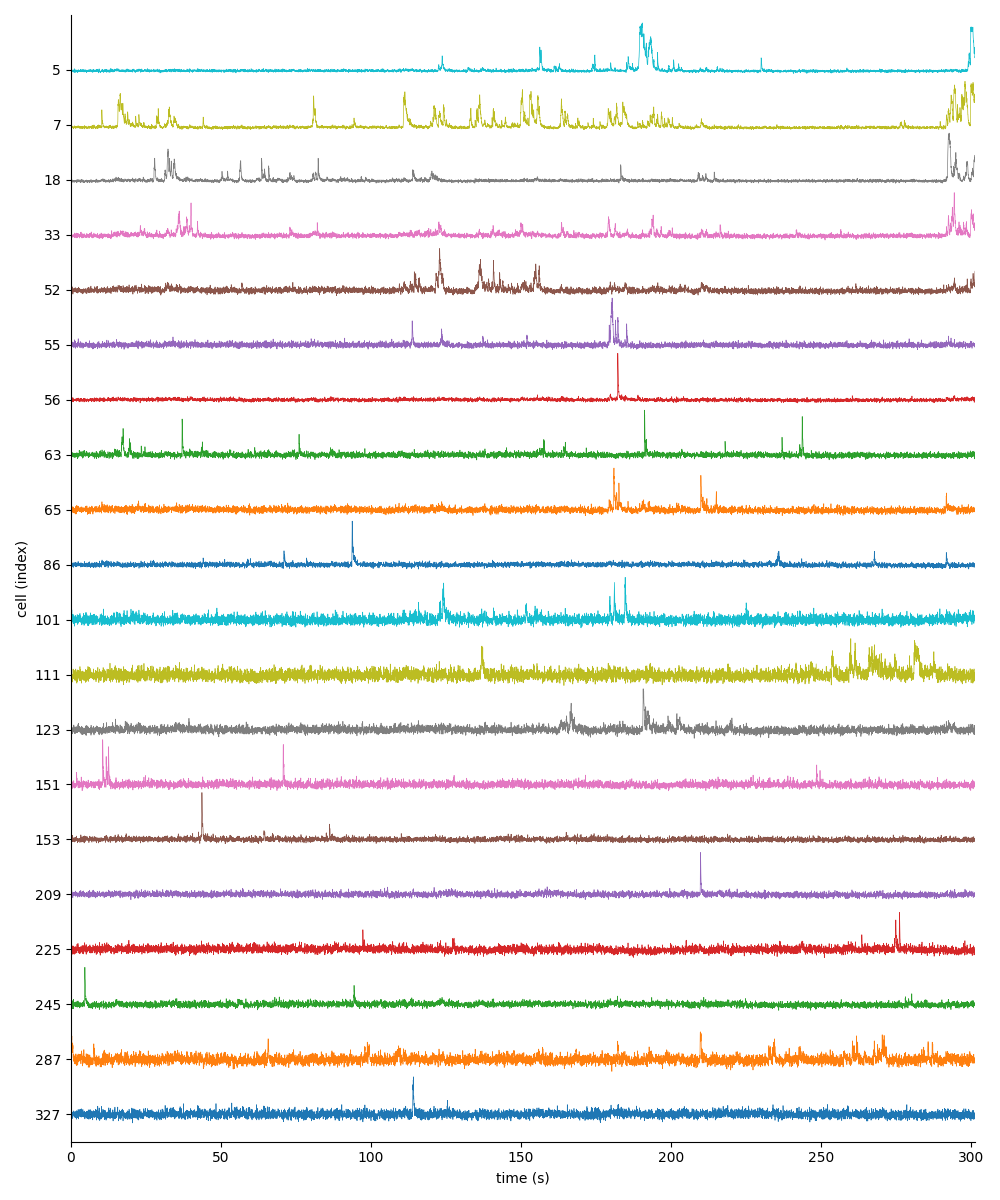 |
 |