Run suite2p pipeline¶
This tool uses 1.0 compute credits per hour.
Overview¶
suite2p is a widely used toolbox for processing 2-photon (2P) calcium imaging datasets.
The IDEAS Run suite2p pipeline tool is a wrapper for the end-to-end pipeline suite2p API function run_s2p().
Run suite2p pipeline comprises the following 7 processing steps:
-
Binary conversion: the input movie(s) is (are) converted into a single suite2p binary file (
BinaryRWFileclass, namely). -
Registration (aka motion correction): each frame is aligned to a reference frame via phase correlation, in a rigid and optionally non-rigid way, to correct for motion artifacts.
-
ROI detection: Regions-Of-Interest (ROIs) are detected using computationally efficient, semi-constrained matrix factorization.
-
ROI extraction: neuropil-corrected fluorescence traces and corresponding neuropil fluorescence traces are extracted from the registered movie for all detected ROIs.
-
ROI classification: extracted ROIs, potentially belonging to different cell compartments (cell body aka soma, dendrites, and axon), are curated using either a built-in classifier or a user-provided one, to only keep cell bodies (aka somata) for further analyses.
-
Spike deconvolution: underlying spike events are inferred from the neuropil-corrected fluorescence traces using spike deconvolution based on the OASIS algorithm.
-
Output conversion: the neuropil-corrected fluorescence traces, neuropil fluorescence traces, deconvolved spikes, extraction statistics, classification labels and parameters information can be saved as 4 different formats:
.zipcontaining suite2p's.npyoutputs,.isxdcellset and eventset,.nwband.mat.
Individual tools available
We also provide each of the 7 steps listed above as individual tools.
You can click on any of the step names above to access their own documentation.
Code¶
| Package | Description |
|---|---|
| suite2p fork | To enable working with Inscopix .isxd movies, we forked the official suite2p repository (version 0.14.5.dev1+gbe8830d) and added the ability to directly read .isxd movies. |
| ideas-toolbox-suite2p | This wrapper and its accompanying individual steps are hosted in a Git repository (public release coming soon...). Modules and functions of this package articulate suite2p functions to make them work in the IDEAS ecosystem. |
suite2p documentation¶
For more information and source code from official suite2p sources, follow the links in the table below:
| Link | Description |
|---|---|
| Official documentation | Provides a detailed list of settings (aka parameters), a description of individual steps and a useful FAQ. |
| Git repository | Source code to dive into implementation details. Note that we currently use a fork of this repository, which has the added ability to read Inscopix .isxd files. |
| bioRxiv preprint | "Suite2p: beyond 10,000 neurons with standard two-photon microscopy". Manuscript by Marius Pachitariu, Carsen Stringer, et al. originally posted on the preprint repository bioRxiv in 2016 and updated in 2017, introducing the suite2p toolbox. |
| Curr. Opin. Neurobiol. paper | "Computational processing of neural recordings from calcium imaging data". Peer-reviewed paper by Stringer & Pachitariu, published in Current Opinion in Neurobiology in 2019, and providing many details on the suite2p pipeline. |
| YouTube tutorials | Lectures on registration, cell detection and spike deconvolution, and demo use. |
| OASIS paper | Official paper on the algorithm used for spike deconvolution. |
| J. Neurosci. paper | "Robustness of Spike Deconvolution for Neuronal Calcium Imaging". Peer-reviewed paper by Pachitariu, Stringer and Harris, published in Journal of Neuroscience in 2018, and detailing the suite2p implementation of OASIS. |
Input files¶
| Source Parameter | File Type | File Format |
|---|---|---|
| Raw Movie Files | miniscope_movie, bruker2p_movie, image | isxd, zip, tiff |
| Optional Parameters File | config | npy |
| Optional Classifier File | suite2p_classifier | npy |
As of version 1.0.0, Run suite2p pipeline supports 3 types of input 2P movies:
| Format | Extension(s) | Notes |
|---|---|---|
| Inscopix miniscope movies | .isxd |
Currently, .isxd files can only contain single-plane, single-channel movies. |
| Bruker Ultima2P movies | .zip containing 1 .xml, 1 .env and >=1 .ome.tif files |
Both <5.8 and >=5.8 formats are supported. Also, only single-plane, single-channel movies have been thoroughly tested so far. |
| Standard tiff stack of movie frames | .tiff or .tif |
All .tiff input movies that are supported by suite2p are supported by this IDEAS tool. Refer to the relevant suite2p docs for further information. |
Preparing Bruker Ultima2P movies for upload
For Bruker Ultima2P movies, note that you should zip the parent directory containing the .xml, .env and .ome.tif file(s) (or zip all those files directly), and upload the resulting .zip file to your IDEAS dataset.
Also, please make sure that the .zip file only contains files from the movie you want to process (i.e., do not put several distinct movies in the same .zip file), as all files are extracted in a single, flat directory structure.
Once the .zip file is uploaded on IDEAS, you should set its Type to Bruker 2P Movie in the Files panel, so that it is recognized as a valid input for the suite2p tools on IDEAS.
Using an optional parameters file
We exposed a curated list of the most impactful parameters (listed in the next section) in the IDEAS Analysis Table.
However, if you want a more granular control on all of the parameters exposed in suite2p's run_s2p(), you can upload an optional .npy parameters file as an input of Run suite2p pipeline.
Note that for parameters that are present both in this optional .npy parameters file and in the IDEAS Analysis Table, you can choose which source has priority for setting those parameters' values by using Get Parameters Values from in the Main Settings of the IDEAS Analysis Table.
You can download here .npy parameters files for default 2P and 1P-optimized parameters.
To modify parameter values in a .npy parameters file, you can use the following code:
import numpy as np
ops = np.load(<path_to_input_ops_npy>, allow_pickle=True).item()
ops[<parameter_name>] = <parameter_value>
np.save(<path_to_output_ops_npy>, ops)
For example, to set high_pass to 200 in ops.npy:
import numpy as np
ops = np.load("ops.npy", allow_pickle=True).item()
ops["high_pass"] = 200
np.save("ops_modified.npy", ops)
Using an optional classifier file
If you prefer using your own cell classifier in place of suite2p's builtin classifier, you can upload an optional .npy classifier file.
It should contain training data as a Python dictionary with 3 keys:
| Key | Description |
|---|---|
stats |
A ROI-by-feature matrix, with for each ROI the values of the selected features (aka predictors) to be used by the classifier, as computed during the ROI detection and ROI extraction steps. |
keys |
A list of feature names, corresponding to the keys of the stats dictionary produced by the ROI detection and extraction steps. |
iscell |
A ROI-length vector containing the accept/reject (or cell/non-cell) labels (aka target), to be used as ground truth by the classifier. |
As an example, the default built-in suite2p classifier contains training data for 14058 ROIs and 3 features ("skew", "compact" and "npix_norm"), and thus has a stats matrix of shape (14058, 3), a keys list as ['skew', 'compact', 'npix_norm'], and an iscell vector of shape (14058,) containing 0 or 1 values as float32 type.
Parameters¶
| Parameter | suite2p name | Required? | Default | Description |
|---|---|---|---|---|
| Raw Movie Files | - | True | N/A | Input 2P movie(s) [.isxd, .zip, .tiff] |
| Optional Parameters File | - | False | N/A | Optional parameters file that allows for more granular control of all available suite2p parameters. See documentation for more details [.npy] |
| Optional Classifier File | classifier_path | False | N/A | [from suite2p docs] "Path to classifier file you want to use for cell classification." [.npy] |
| Overwrite Analysis Table Parameters | - | True | False | When a parameters file is provided as input, whether the parameters from the analysis table columns should be overwritten by the values from the file. Only effective when an optional parameters file is provided. Defaults to False. [bool] |
| Tau | tau | False | 1.0 | [from suite2p docs] "The timescale of the sensor (in seconds), used for deconvolution kernel. The kernel is fixed to have this decay and is not fit to the data. We recommend: 0.7 for GCaMP6f; 1.0 for GCaMP6m; 1.25-1.5 for GCaMP6s." [float, =0.001] |
| Frames to Include | frames_include | False | -1 | [from suite2p docs] "If greater than zero, only [the first] <frames_include frames are processed. Useful for testing parameters on a subset of data." Note that suite2p will not run on less than 50 frames, and might have unpredictable behavior on less than 200 frames [int, -1 or =50] |
| Save NPY | - | False | True | If true, save suite2p NPY output (as a ZIP file) [bool] |
| Save ISXD | - | False | True | If true, save suite2p output as ISXD files [bool] |
| Save NWB | save_NWB | False | False | [from suite2p docs] "Whether to save output as NWB file." [bool] |
| Save MAT | save_mat | False | False | [from suite2p docs] "Whether to save the results in matlab format in file "Fall.mat"." [bool] |
| Maximum Registration Shift | maxregshift | False | 0.1 | [from suite2p docs] "The maximum shift as a fraction of the frame size. If the frame is Ly pixels x Lx pixels, then the maximum pixel shift in pixels will be max(Ly,Lx) * ops['maxregshift']." [float, [0.01 1]] |
| Threshold for Bad Frames | th_badframes | False | 1.0 | [from suite2p docs] "Involved with setting threshold for excluding frames for cropping. Set this smaller to exclude more frames." [float, =0] |
| Nonrigid Registration | nonrigid | False | True | [from suite2p docs] "Whether or not to perform non-rigid registration, which splits the field of view into blocks and computes registration offsets in each block separately." [bool] |
| ROI Threshold Scaling | threshold_scaling | False | 1.0 | [from suite2p docs] "This controls the threshold at which to detect ROIs (how much the ROIs have to stand out from the noise to be detected). If you set this higher, then fewer ROIs will be detected, and if you set it lower, more ROIs will be detected." [float, =0] |
| Neuropil Coefficient | neucoeff | False | 0.7 | [from suite2p docs] "Neuropil coefficient for all ROIs." [float, =0] |
| Spikes Percentile Threshold | - | False | 99.7 | Threshold for denoising the deconvolved spike trains, in percentile. Any value in the ROIs-by-time-point deconvolved spike matrix that is below the matrix's xth percentile value is set to 0. Note that the same threshold is applied to all ROIs, and that this thresholding step does not binarize the deconvolved spike trains but simply filters out the low-amplitude spike events [float, [0 100)] |
| FOV vmin Percentile | - | False | 0 | Minimum value for the colormap range, as percentile of the FOV fluorescence [float, [0 |
| FOV vmax Percentile | - | False | 99 | Maximum value for the colormap range, as percentile of the FOV fluorescence [float, ( |
| FOV Colormap | - | False | plasma | Colormap for plotting the FOV [str] |
| Show Grid on FOV | - | False | True | Whether or not to show the grid on FOVs [bool] |
| FOV Ticks Step | - | False | 128 | Step for the x- and y-ticks [int, >0] |
| Movie Display Rate | - | False | 10 | Display rate for the preview movies, in Hz [float, >=0.1] |
| Number of Sample Cells | - | False | 20 | Number of sample cells for the cell extraction preview [int, >0] |
| Random Seed | - | False | 0 | Random seed for selecting sample cells [int, >=0] |
| Show Non-Sample Cells' Footprints | - | False | True | Whether or not to show footprints of non-sample cells on the cell footprint FOV image. If False, only footprints of sample cells are displayed [bool] |
Frames to Include
While technically frames_include (aka Frames to Include in the IDEAS Analysis Table) can be set to either -1 (use all frames) or any value >0, suite2p does not run on less than 51 frames, and it warns that "unpredictable behaviors may occur" if less than 201 frames are provided (see code here, L289-290 and L291-294, respectively). Therefore, we recommend setting Frames to Include to either -1 or any value >200.
Automatically set parameters
The following parameters are set to values automatically detected in the input file's metadata: nplanes, nchannels, and fs. However, you can set those parameters to a value of your choice by using an optional .npy parameters file.
Hard-coded parameters
The following few parameters are hard-coded to guarantee code integrity: force_sktiff, multiplane_parallel, fast_disk, delete_bin, bruker, nwb_driver, save_path0, save_folder, look_one_level_down, subfolders, move_bin, combined, keep_movie_raw, reg_tif, reg_tif_chan2, and roidetect. There parameters need to be hard-coded to ensure the code works in the IDEAS cloud environment, specifically to prevent writing permission or file not found errors. Therefore, even changing these parameters in a .npy parameters file will not cause any effect and will be overridden by the IDEAS hard-coded values. Note that these hard-coded parameters do not affect the data processing logic of suite2p, and are only relevant to set read/write permissions on the computer where suite2p is being executed.
Algorithm overview¶
Run suite2p pipeline processes 2P data through 7 steps illustrated in the flowchart below:
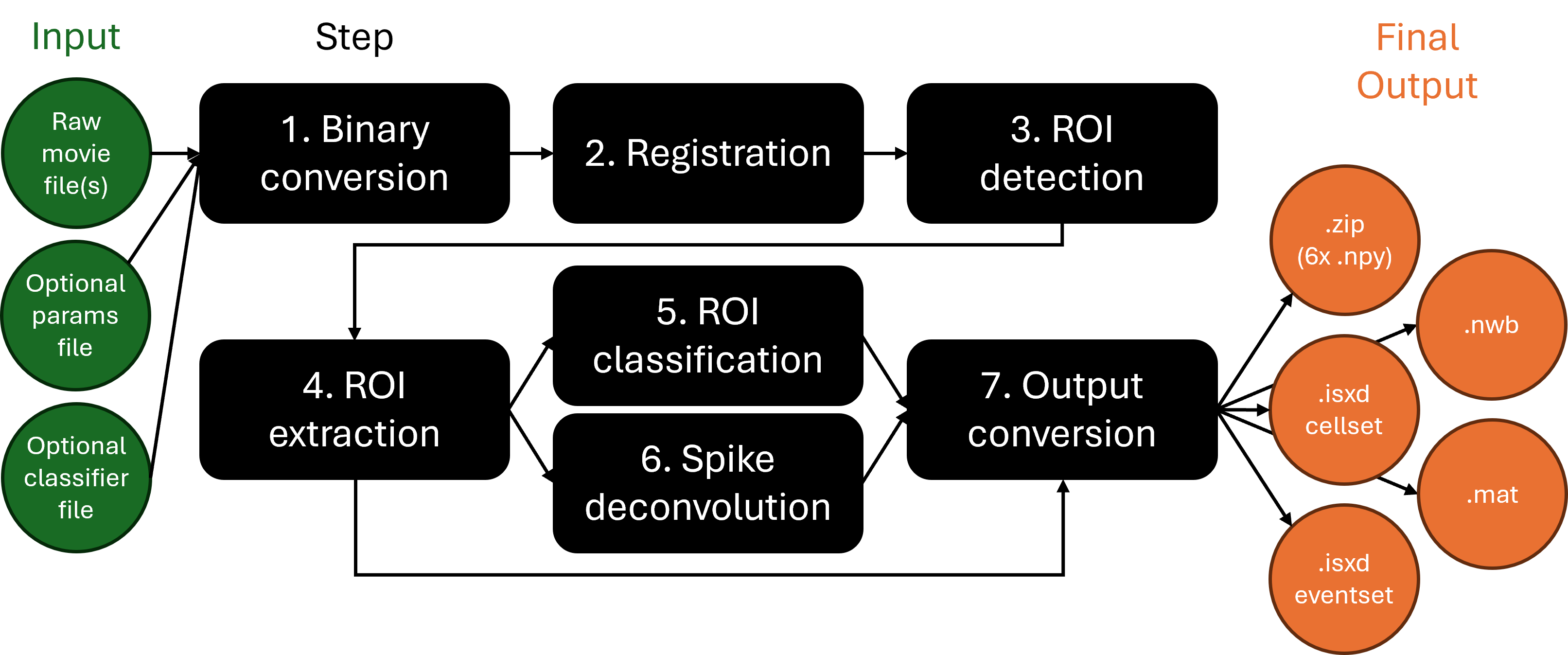
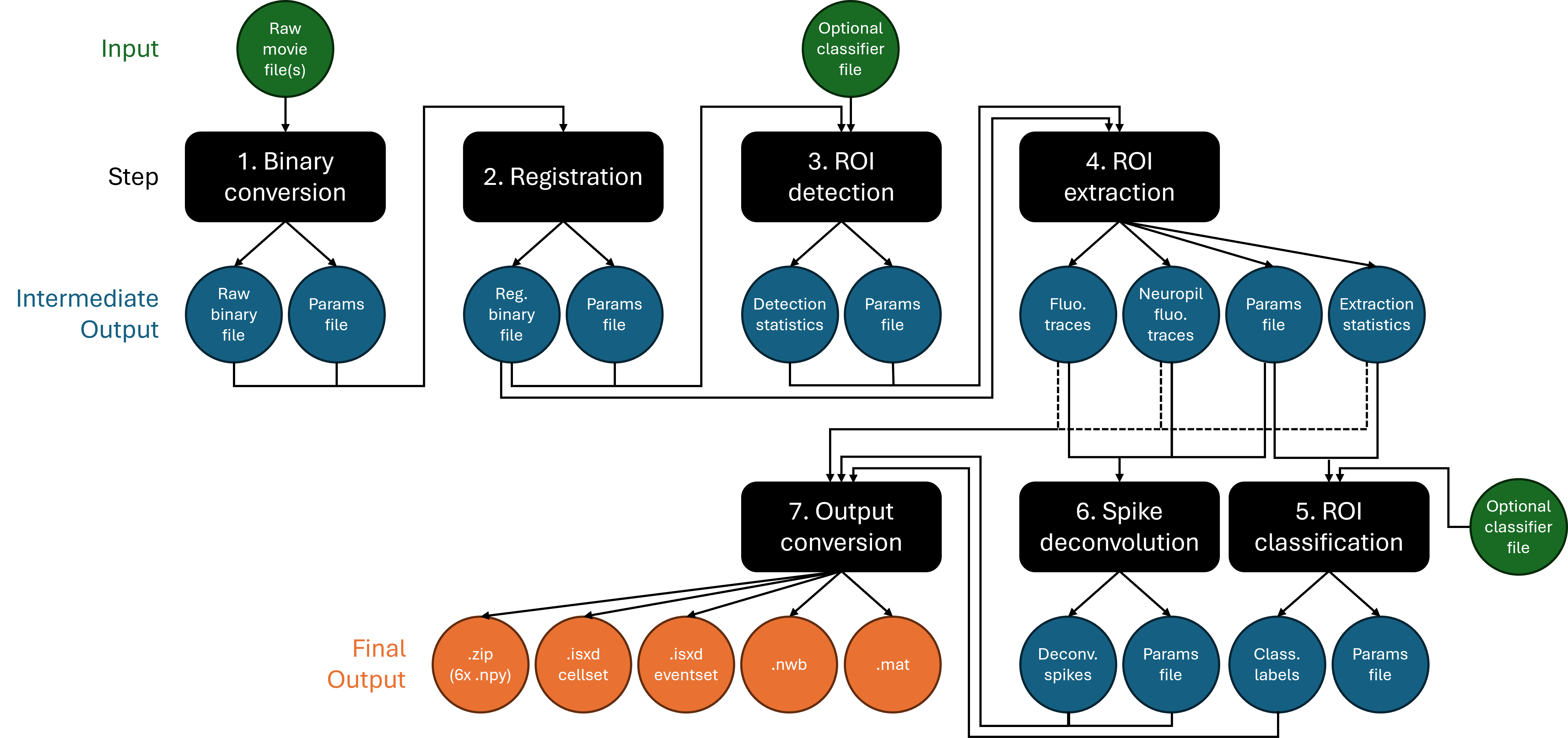
Note that you can also achieve the same workflow by executing the invididual steps separately. This approach offers more modularity and exposure to all parameters in the IDEAS Analysis Tables. In this case, the flow chart looks like:
Output files¶
You can choose the format(s) of Run suite2p pipeline's output among 4 distinct, non-mutually exclusive options:
| Format | Extension(s) | Notes |
|---|---|---|
| suite2p output | .zip file containing 6 .npy files |
As listed in the suite2p documentation. |
| Inscopix cellset and eventset | 2 separate .isxd files |
Compatible with secondary analysis tools on IDEAS. |
| Neurodata Without Borders | .nwb |
Note that suite2p .nwb outputs are not compatible with DANDI out of the box. To create DANDI-compatible suite2p outputs, you can use the IDEAS Convert to NWB tool. |
| MATLAB | .mat |
For further processing or secondary analyses in MATLAB. |
Each output file has metadata and previews attached to it.
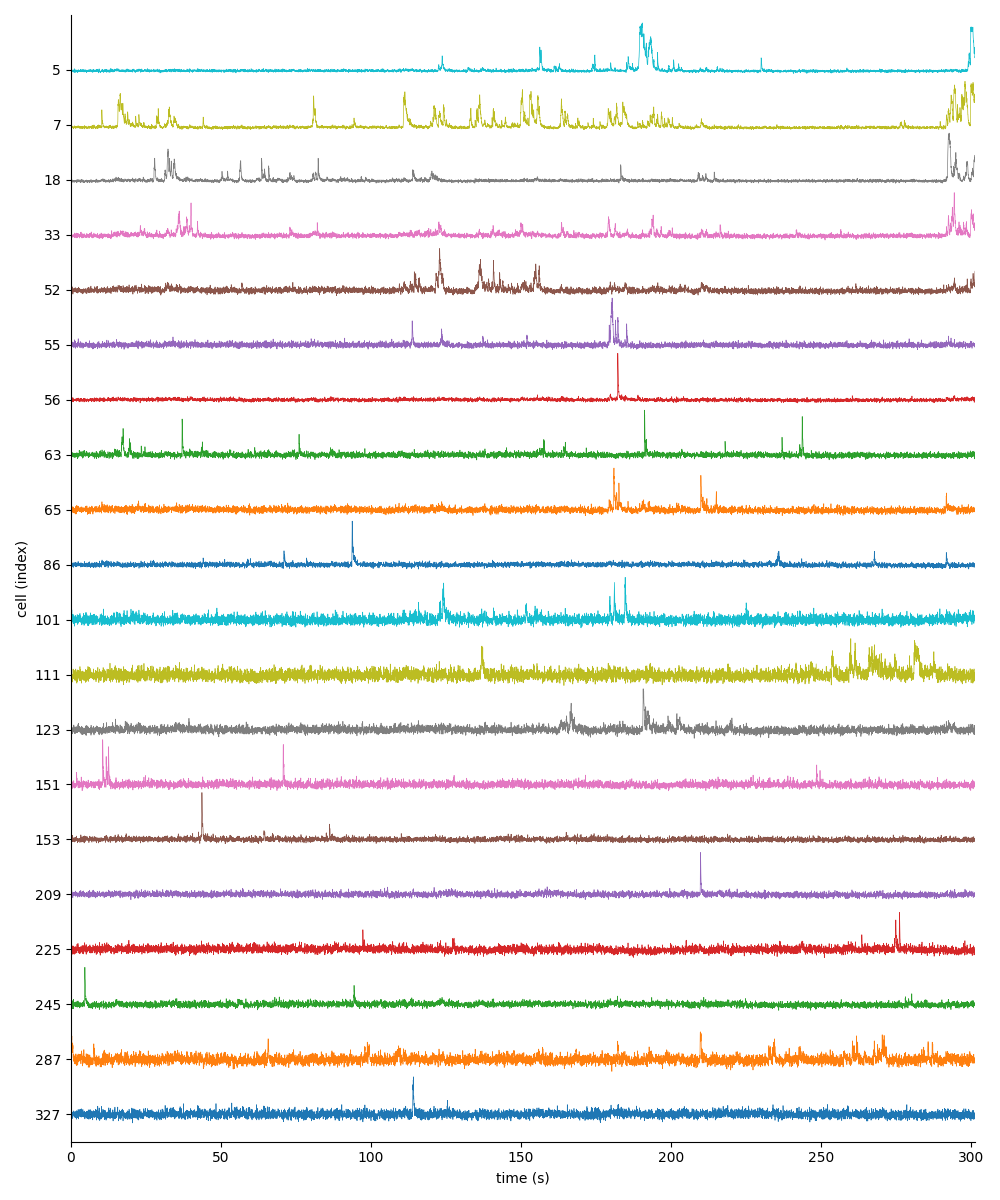
For suite2p .zip, .nwb and .mat outputs, you can explore 8 preview figures and 1 preview movie, illustrating various aspects of the processing pipeline.
You can find below an example of these previews obtained from processing a 5-minute 2P movie of mouse cortex (data courtesy of Dr. Ahmet Arac, MD, at UCLA):
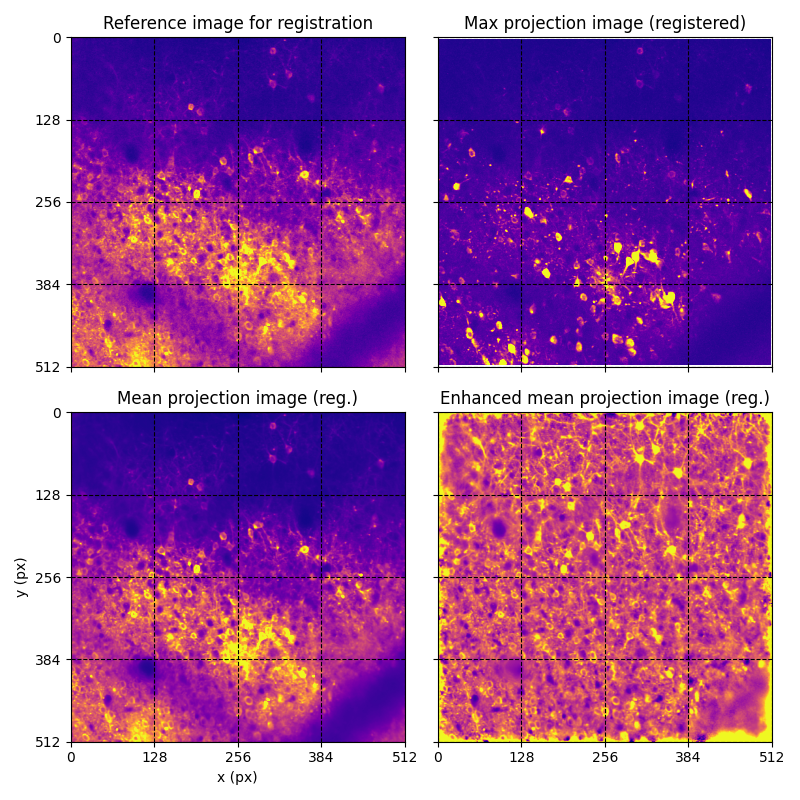 |
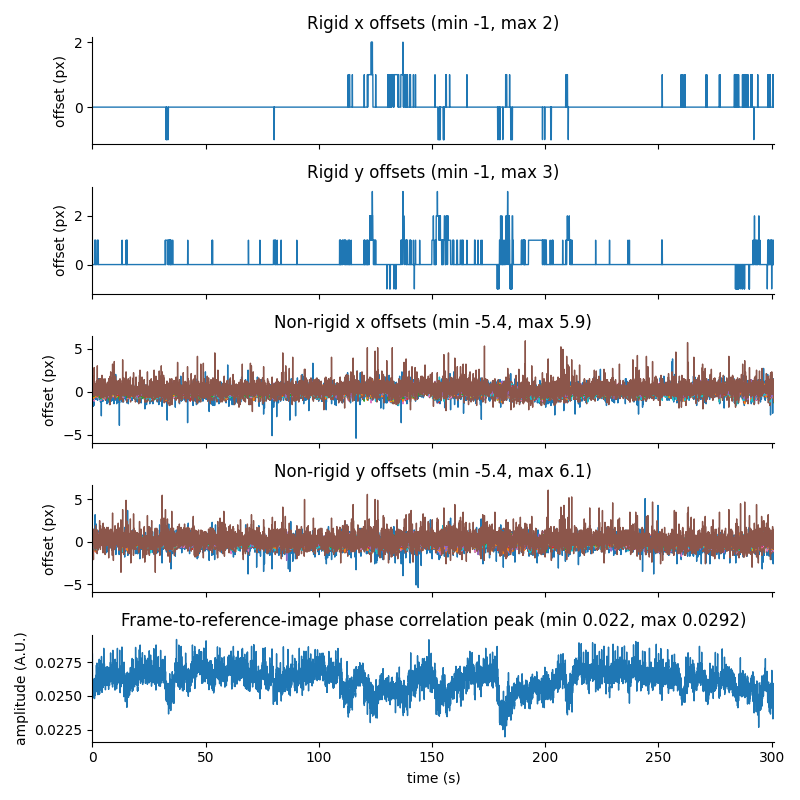 |
|
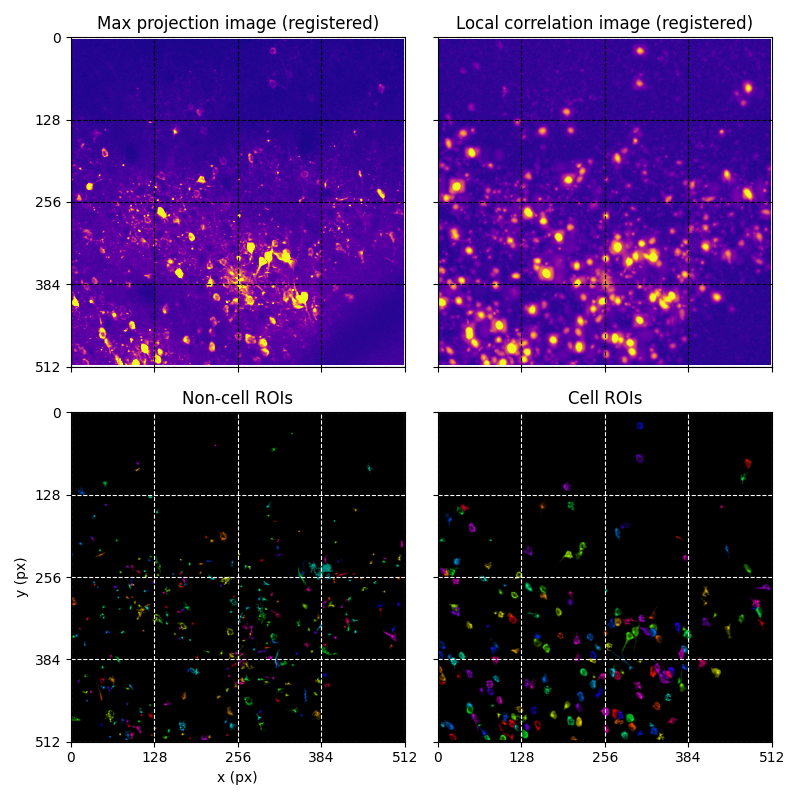 |
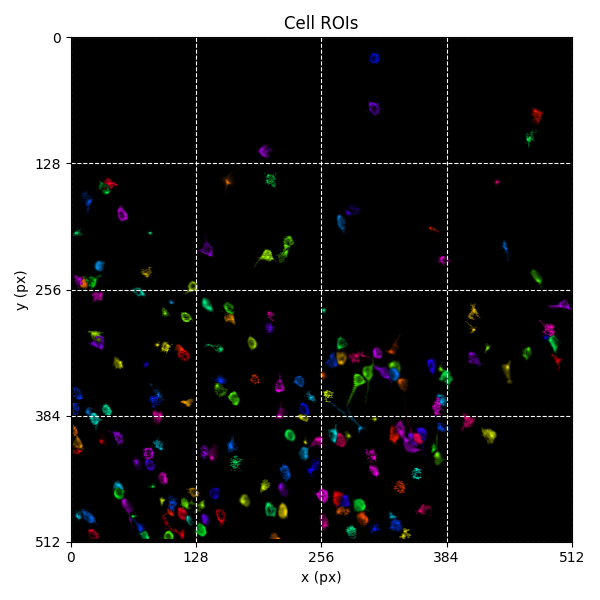 |
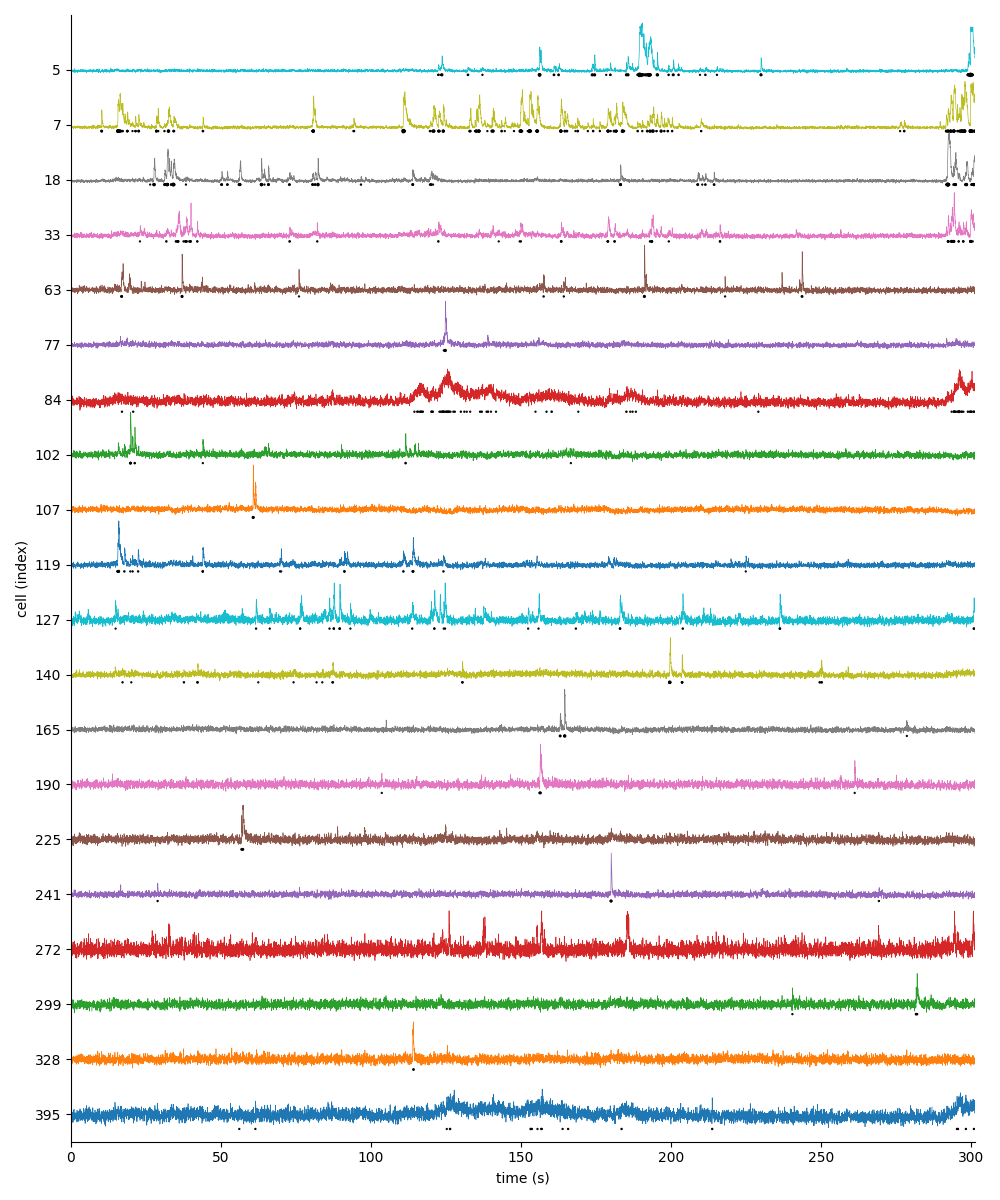 |
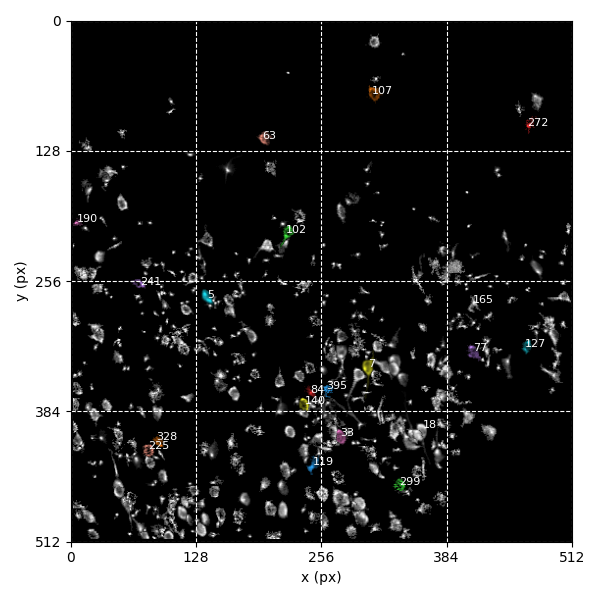 |
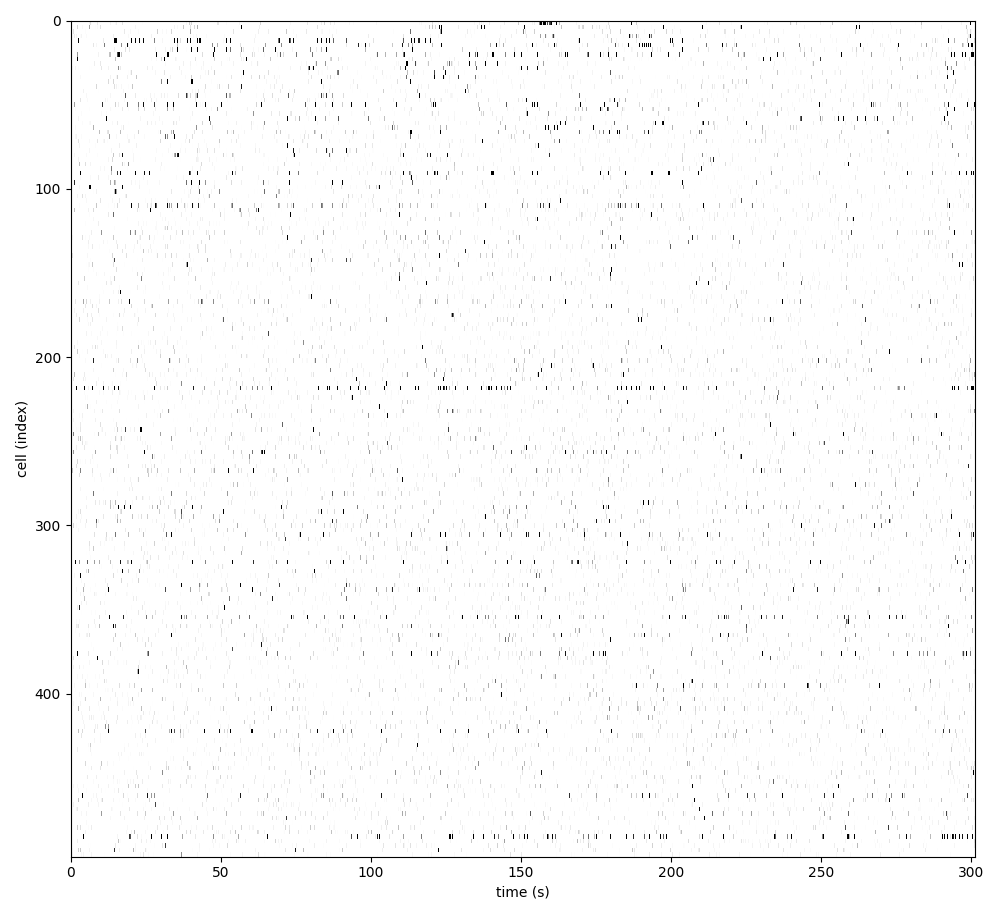 |
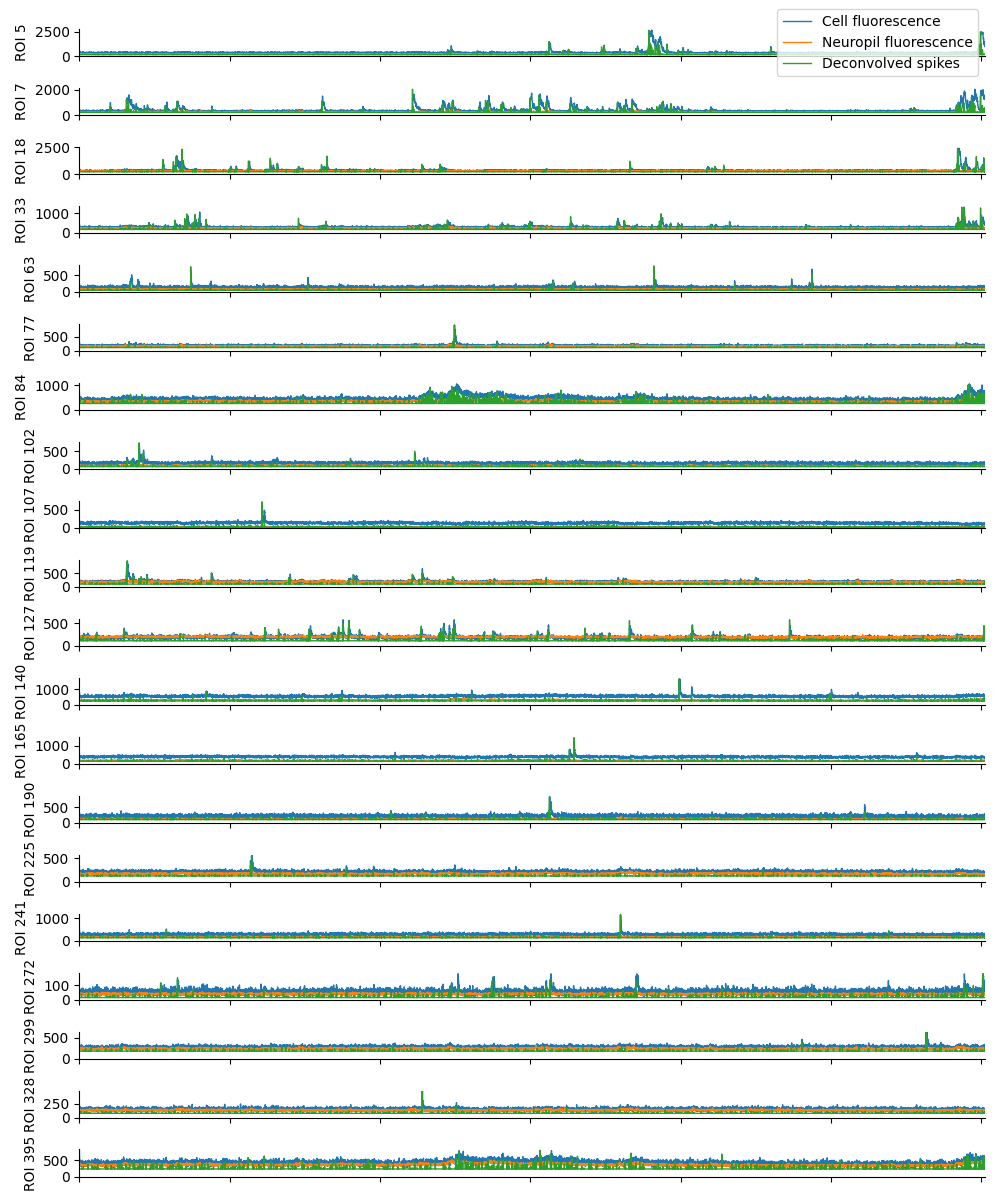 |
For Inscopix .isxd outputs, you can explore 2 preview figures for cellset_raw.isxd:
 |
 |
and 2 for eventset.isxd (also obtained from processing the same 5-minute 2P movie as above):
 |
 |
Modifying output figures
We provide a set of visualization parameters in the Analysis Table for controlling several graphical aspects of the preview figures (listed above; e.g., colormap, number of sample cells, etc), so you can get as close to presentation or publication-ready rendering.
Next steps¶
If you choose to output to .isxd, there are many compatible downstream IDEAS tools for further processing and analysis, including combining with other experimental and behavioral data:
-
Data quality evaluation and cell filtering
-
Analysis
FAQ¶
| Question | Answer |
|---|---|
| Will this tool work on non-Bruker 2P movies? | Besides Bruker 2P movies, we currently also support Inscopix .isxd movies and .tif/.tiff standard format stack of frames. If you are interested in using suite2p on IDEAS with other 2P movie formats, please don't hesitate to reach out to our team! |
| Will this tool work on Inscopix .isxd 1P movies? | Yes, there are actually several parameters that you can set using an optional .npy file to optimize processing of 1P data. In Input files's "Using an optional parameters file" info box, we provide an example 1P-optimized parameters file. |
How can I create my own ops.npy parameters file? |
You can download the default 2P parameters file we provide in Input files's "Using an optional parameters file" info box, open it in Python with numpy using the following line of code: ops = np.load("ops.npy", allow_pickle=True).item(), manually change the parameters of your choice, and save the updated dictionary using the following line of code: np.save("ops_custom.npy", ops). |
| What is the difference between running suite2p on IDEAS versus locally on my own computer? | Besides the general advantages of using IDEAS for storing your data and accessing high-performance computing resources in seamless ways, Run suite2p pipeline applies the exact same processing steps than the official suite2p run_s2p() function, but goes beyond this by offering an end-to-end, code-free experience, compatibility with Inscopix .isxd miniscope movies, and metadata and preview figures for the output files. |