Combine and Compare Peri-Event Analysis Data¶
This tool uses 1.0 compute credits per hour.
Overview¶
This tool is used to combine peri-event analysis data from multiple recordings and perform a statistical comparison between two groups to determine if there is a difference in the fractions of up- or down-modulated neurons.
Parameters¶
| Parameter | Required? | Default | Description |
|---|---|---|---|
| Peri-Event Traces Files | True | N/A | Select peri-event traces from the first group to use for analysis |
| Peri-Event Statistics Files | True | N/A | Select peri-event statistics from the first group to use for analysis |
| Name | True | group1 | Name of the first group |
| Peri-Event Traces Files | False | N/A | Select peri-event traces from the second group to use for analysis |
| Peri-Event Statistics Files | False | N/A | Select peri-event statistics from the second group to use for analysis |
| Name | False | N/A | Name of the second group |
| Comparison Type | False | N/A | Type of statistical test to perform |
| Data Pairing | False | N/A | Indicates whether observations should be paired for statistical comparison |
| Significance Threshold | False | N/A | p-value threshold for classifying neurons as up- or down-modulated |
| Average Method | True | neurons | Specifies how to average the data in each input group. If 'By Neurons' is selected, the neurons from all recordings in the group will be pooled together and averaged to obtain the group average. If 'By Recordings' is selected, the data contained in each recording will be averaged independently, then the recordings will be averaged together to obtain the group average. |
| Tolerance | True | 0.01 | Maximum time shift in seconds between the time windows of the input traces files |
| Population activity (y-axis range) | False | auto | y-axis range (z-score) applied to the event-aligned population activity plot specified as 'min,max' (e.g. -1,1) or 'auto' |
| Activity heatmap (colormap range) | False | auto | Colormap range (z-score) applied to the activity heatmap specified as 'min,max' (e.g. -1,1) or 'auto' |
| Activity by modulation group (y-axis range) | False | auto | y-axis range (z-score) applied to the event-aligned activity by modulation plot specified as 'min,max' (e.g. -1,1) or 'auto' |
| Group colors | False | #1f77b4, #ff7f0e | Colors that represent group 1 and 2 respectively |
| Modulation colors | False | green,blue,black | Colors that represent up-, down- and non-modulation respectively |
| Activity heatmap colormap | False | coolwarm | Colormap applied to the activity heatmap |
Valid Inputs¶
| Source Parameter | File Type | File Format |
|---|---|---|
| Peri-Event Traces Files | neural_traces, event_aligned_neural_data | csv, csv |
| Peri-Event Statistics Files | statistics | csv |
| Peri-Event Traces Files | neural_traces, event_aligned_neural_data | csv, csv |
| Peri-Event Statistics Files | statistics | csv |
Note
Only Matplotlib-compatible colors are supported for Group colors and Modulation colors, and Matplotlib-compatible colormaps are supported for Activity Heatmap colormap. Invalid color values will revert to the default settings shown in the Parameters table.
Algorithm Description¶
The workflow is summarized using a flowchart below.
The workflow begins by combining the data in the first input group. If no data is selected for the second input group, the workflow terminates upon combining the data from the first group. If data is selected for the second group, that data will be combined independently of the first group. Finally, the two groups are compared to determine if there is statistically significant difference between them.
Combination¶
The combination step is performed independently for each input group. The process starts with the alignment of time windows across the input files. If the difference between the time windows is within the specified tolerance, the algorithm will align the time points across the input files and trim off excess time points. The data from all recordings in the group is then aggregated, creating a population of cells. Individual cells are classified as up-, down-, or non-modulated based on the significance threshold parameter. The fraction of up-, down-, and non-modulated neurons across the aggregated population of cells is computed. The event-aligned population activity is obtained by averaging the neural activity at each time point. When averaging across neurons, neurons from all recordings in the group are pooled together and averaged to obtain the group average. When averaging across recordings, the data is first averaged across neurons for each recording, yielding a single average trace per recording. The recording traces are then averaged together to obtain the group average. Likewise, the event-aligned sub-population activity is obtained for each modulation category (up-modulated, down-modulated, non-modulated) by averaging the neural activity of the corresponding cells.
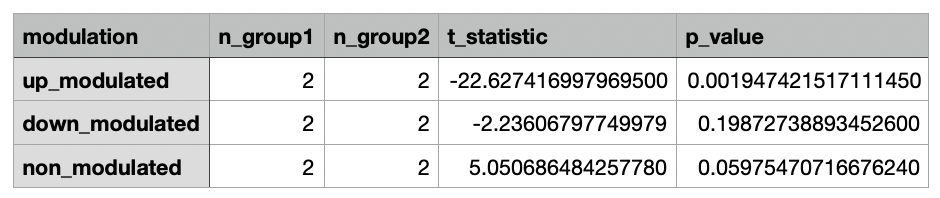
Comparison¶
The fraction of up-, down-, and non-modulated neurons is computed for each recording. A t-test is then performed to compare the means of the two groups for each modulation category (up-modulated, down-modulated, non-modulated). The t-statistic and corresponding p-value is reported for each statistical test. We specifically test for the null hypothesis that the two groups have identical average values. The alternative hypothesis tested is determined by the comparison type parameter as described in the table below.
Modulation significance and p-values¶
- Per-cell p-values come from the Peri-Event Analysis Workflow, which circularly permutes event times to build a null distribution. For each cell, the workflow records
p_value = min(p_high, p_low)wherep_highis the fraction of null Δ values greater than observed andp_lowis the fraction less than observed (in[0, 0.5], resolution1 / Number of random shuffles). - When combining, cells are reclassified using the selected
Significance threshold (α)with one-tailed criteria atα/2to implement a two-tailed decision: up-modulated ifp_high < α/2, down-modulated ifp_low < α/2, otherwise non-significant. No new permutations are run during combination. - Group comparisons operate on the resulting modulation fractions and use t-tests; the alternative hypothesis is controlled by
Comparison type, and pairing is controlled byData pairing.
| Comparison Type | Hypothesis Tested |
|---|---|
| One-Tailed (Less) | fraction of modulated neurons in the first group is less than the second group |
| One-Tailed (Greater) | fraction of modulated neurons in the first group is greater than the second group |
| Two-Tailed (Unequal) | fraction of modulated neurons in the first group is different from the second group, i.e. fraction of modulated neurons in the the first group is either less than or greater than the second group |
Output Figures¶
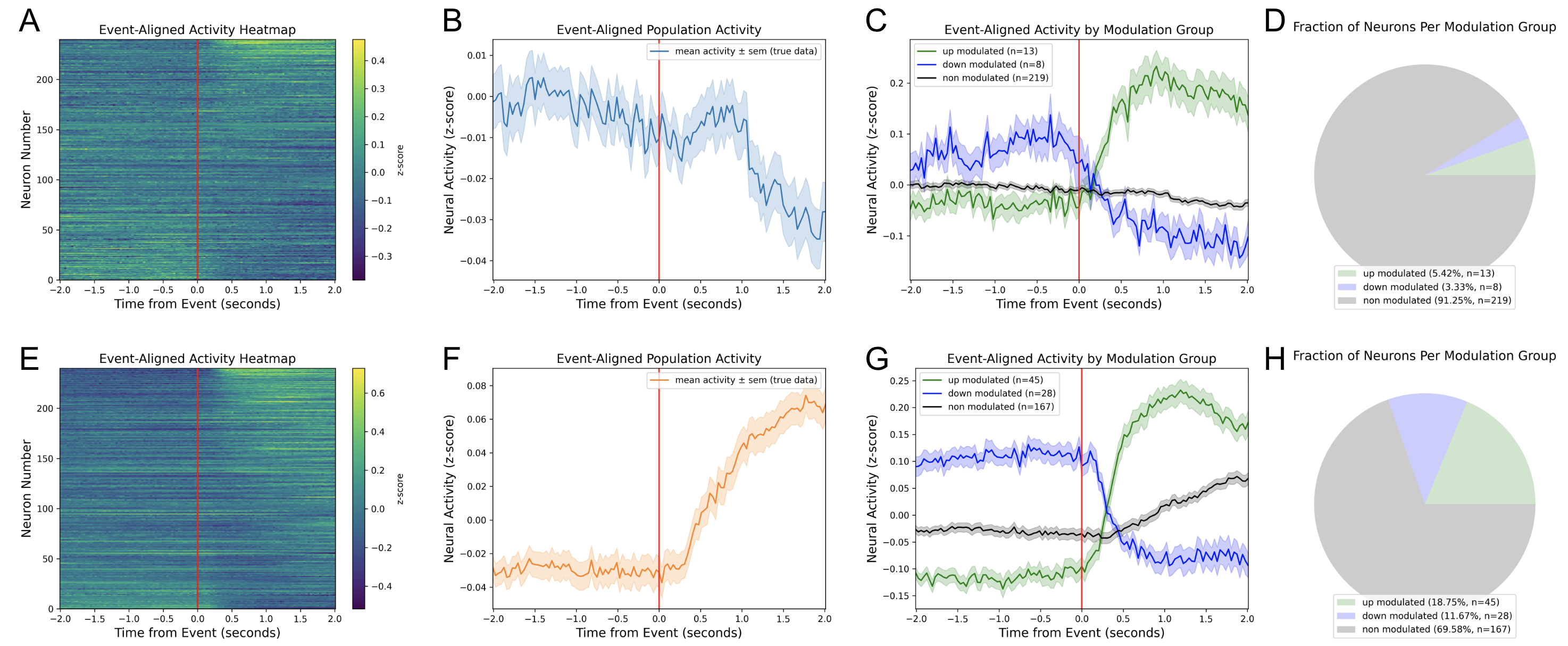
Combination Figures¶
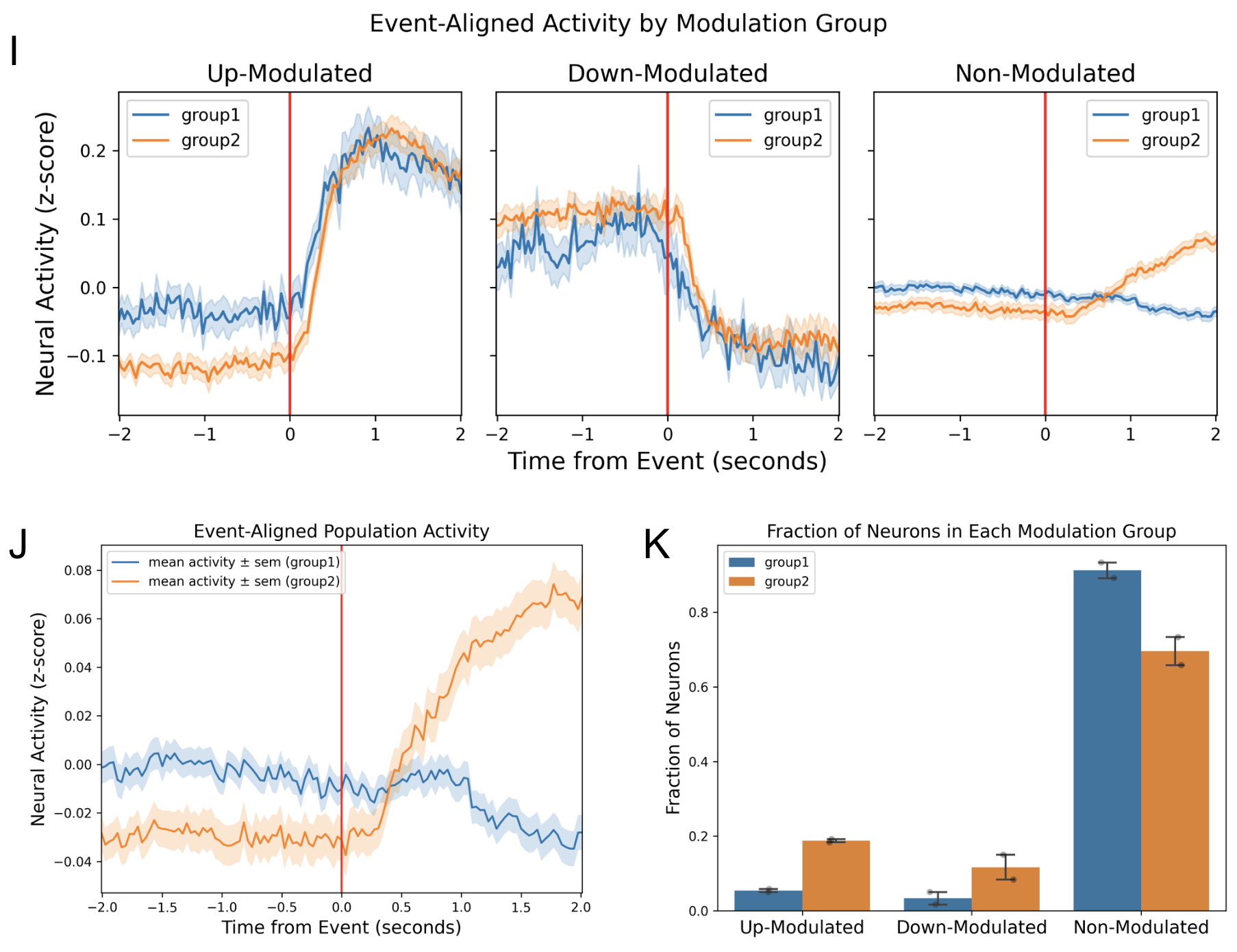
Comparison Figures¶
Legend¶
| Panel | Description |
|---|---|
| A, E | Event-aligned activity heatmaps of the first and second groups, respectively. |
| B, F | Event-aligned population activity of the first and second groups, respectively. |
| C, G | Event-aligned sub-population activity of the first and second groups, respectively. |
| D, H | Pie chart showing the fraction of up-, down-, and non-modulated neurons in the first and second groups, respectively. |
| I | Comparison of event-aligned sub-population activity between the two input groups. |
| J | Comparison of event-aligned population activity between the two input groups. |
| K | Comparison of the fraction of neurons in each modulation category between the two input groups. The mean ± sem is plotted along with the individual data points, each representing a recording session. |
Output Data¶
Combination Data¶
For each input group, two files are produced during the combination step. The two files are equivalent to those produced by the peri-event analysis workflow.
Event-Aligned Traces¶
A csv file containing the event-aligned activity of all cells as well as the activity of the population and sub-populations.
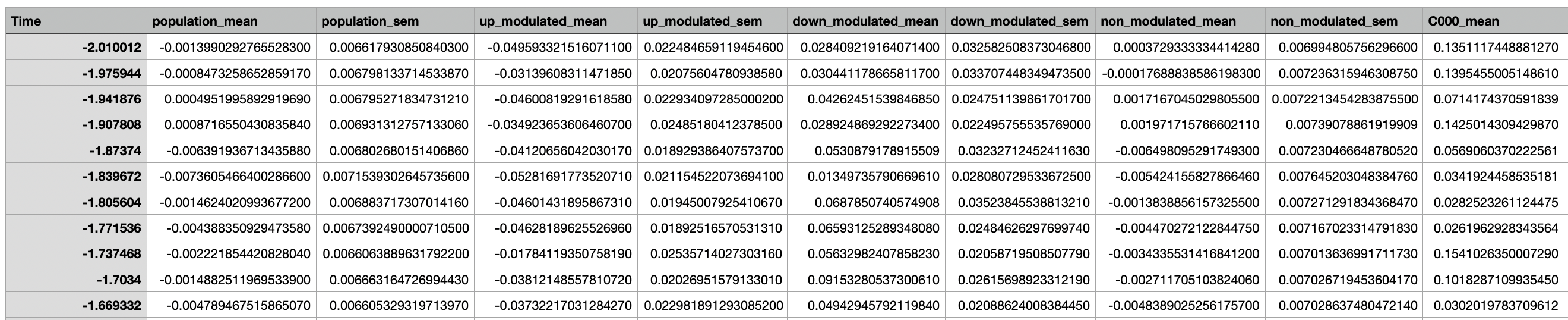
Event-Aligned Statistics¶
A csv file containing the data used to perform statistical tests for each cell along with corresponding
statistics and modulation classification.
Comparison Data¶
A single csv file containing a summary of the statistical tests performed.