suite2p output conversion¶
This tool uses 0.5 compute credits per hour.
Overview¶
This suite2p output conversion tool is a wrapper for the suite2p functions io.save_nwb() and io.save_mat(), as well as for zipping the .npy output files and using the npy_to_isxd() function from our ideas-toolbox-suite2p package.
You can find more information on this processing step in the suite2p docs.
suite2p output conversion saves the neuropil-corrected fluorescence traces, neuropil fluorescence traces, deconvolved spikes, extraction statistics, classification labels and parameters information as 4 different formats: .zip containing suite2p's .npy outputs, .isxd cellset and eventset, .nwb and .mat.
Note that this tool constitutes the first step of Run suite2p pipeline.
Input files¶
| Source Parameter | File Type | File Format |
|---|---|---|
| Fluorescence Traces File | suite2p_data | npy |
| Neuropil Fluo File | suite2p_data | npy |
| Deconvolved Spikes File | suite2p_data | npy |
| Extraction Statistics File | suite2p_data | npy |
| Parameters File | config | npy |
| Classification Labels File | suite2p_data | npy |
Parameters¶
| Parameter | suite2p name | Required? | Default | Description |
|---|---|---|---|---|
| Fluorescence Traces File | - | True | N/A | Input fluorescence traces file, as outputted by the ROI extraction step [ |
| Neuropil Fluo File | - | True | N/A | Input neuropil fluorescence traces file, as outputted by the ROI extraction step [ |
| Deconvolved Spikes File | - | True | N/A | Input deconvolved spikes file, as outputted by the spike deconvolution step [ |
| Extraction Statistics File | - | True | N/A | Input extraction statistics file, as outputted by the ROI extraction step [ |
| Parameters File | - | True | N/A | Input parameters file, as outputted by the spike deconvolution step [ |
| Classification Labels File | - | True | N/A | Input classification labels file, as outputted by the ROI classification step [ |
| Save NPY | - | False | True | If true, save suite2p NPY output (as a ZIP file) [bool] |
| Save ISXD | - | False | True | If true, save suite2p output as ISXD files [bool] |
| Save NWB | save_NWB | False | False | [from suite2p docs] "Whether to save output as NWB file." [bool] |
| Save MAT | save_mat | False | False | [from suite2p docs] "Whether to save the results in matlab format in file "Fall.mat"." [bool] |
| Spikes Percentile Threshold | - | False | 99.7 | Threshold for denoising the deconvolved spike trains, in percentile. Any value in the ROIs-by-time-point deconvolved spike matrix that is below the matrix's xth percentile value is set to 0. Note that the same threshold is applied to all ROIs, and that this thresholding step does not binarize the deconvolved spike trains but simply filters out the low-amplitude spike events [float, [0 100)] |
| FOV vmin Percentile | - | False | 0 | Minimum value for the colormap range, as percentile of the FOV fluorescence [float, [0 |
| FOV vmax Percentile | - | False | 99 | Maximum value for the colormap range, as percentile of the FOV fluorescence [float, ( |
| FOV Colormap | - | False | plasma | Colormap for plotting the FOV [str] |
| Show Grid on FOV | - | False | True | Whether or not to show the grid on FOVs [bool] |
| FOV Ticks Step | - | False | 128 | Step for the x- and y-ticks [int, >0] |
| Number of Sample Cells | - | False | 20 | Number of sample cells for the cell extraction preview [int, >0] |
| Random Seed | - | False | 0 | Random seed for selecting sample cells [int, >=0] |
| Show Non-Sample Cells' Footprints | - | False | True | Whether or not to show footprints of non-sample cells on the cell footprint FOV image. If False, only footprints of sample cells are displayed [bool] |
Output files¶
| File name | File type | Notes |
|---|---|---|
| suite2p_output.zip | .zip file |
Contains 6 .npy files outputted by in the previous steps: F.npy, Fneu.npy, spks.npy, stat.npy, iscell.npy, and ops_spike_deconvolution.npy. |
| cellset_raw.isxd | Inscopix .isxd file |
Cell set containing the neuropil-corrected fluorescence traces for all extracted ROIs, as well as ROI footprints and classification labels. |
| eventset.isxd | Inscopix .isxd file |
Event set containing the spike trains for all extracted ROIs. |
| ophys.nwb | .nwb file |
Contains all the same output as suite2p_output.zip but in a .nbw format. |
| Fall.mat | .mat file |
Contains all the same output as suite2p_output.zip but in a .mat format. |
Each output file has metadata and previews attached to it.
For suite2p .zip, .nwb and .mat outputs, you can explore 8 preview figures, illustrating various aspects of the processing pipeline.
You can find below an example of these previews obtained from processing a 5-minute 2P movie of mouse cortex (data courtesy of Dr. Ahmet Arac, MD, at UCLA):
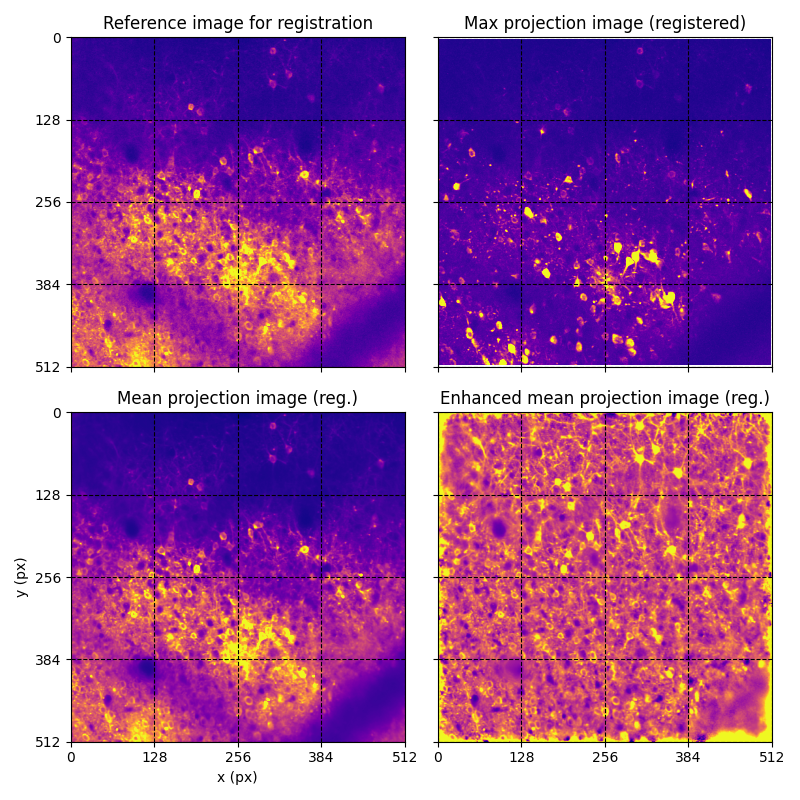 |
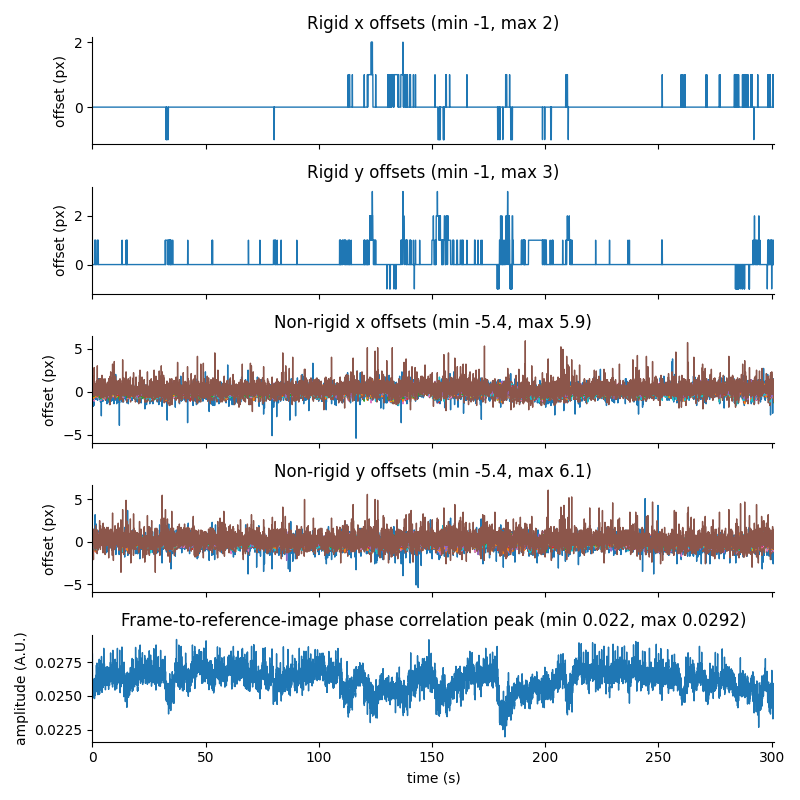 |
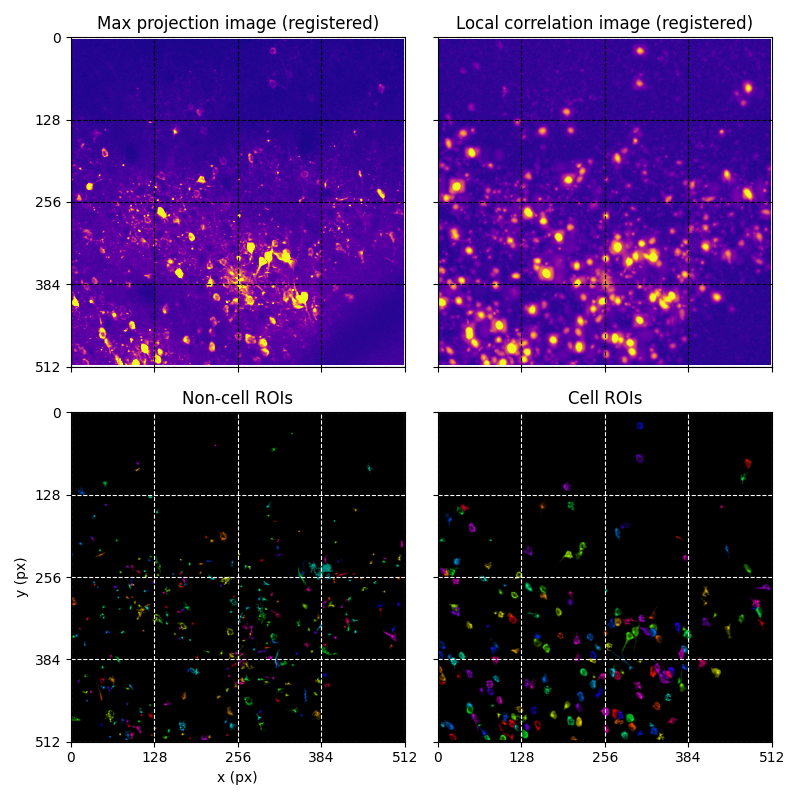 |
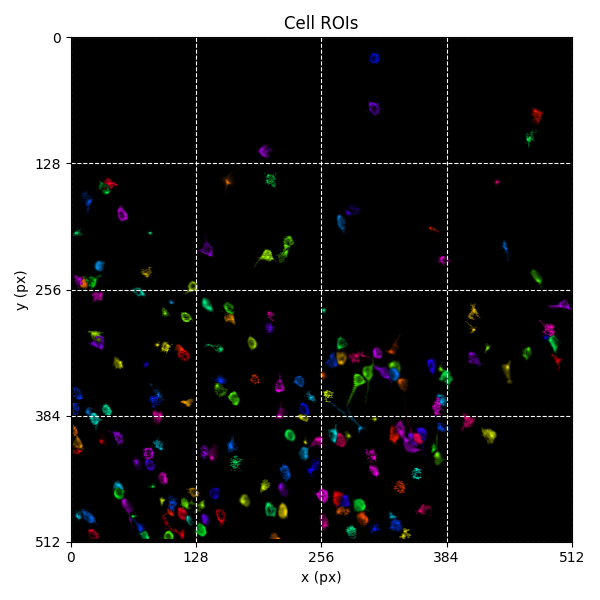 |
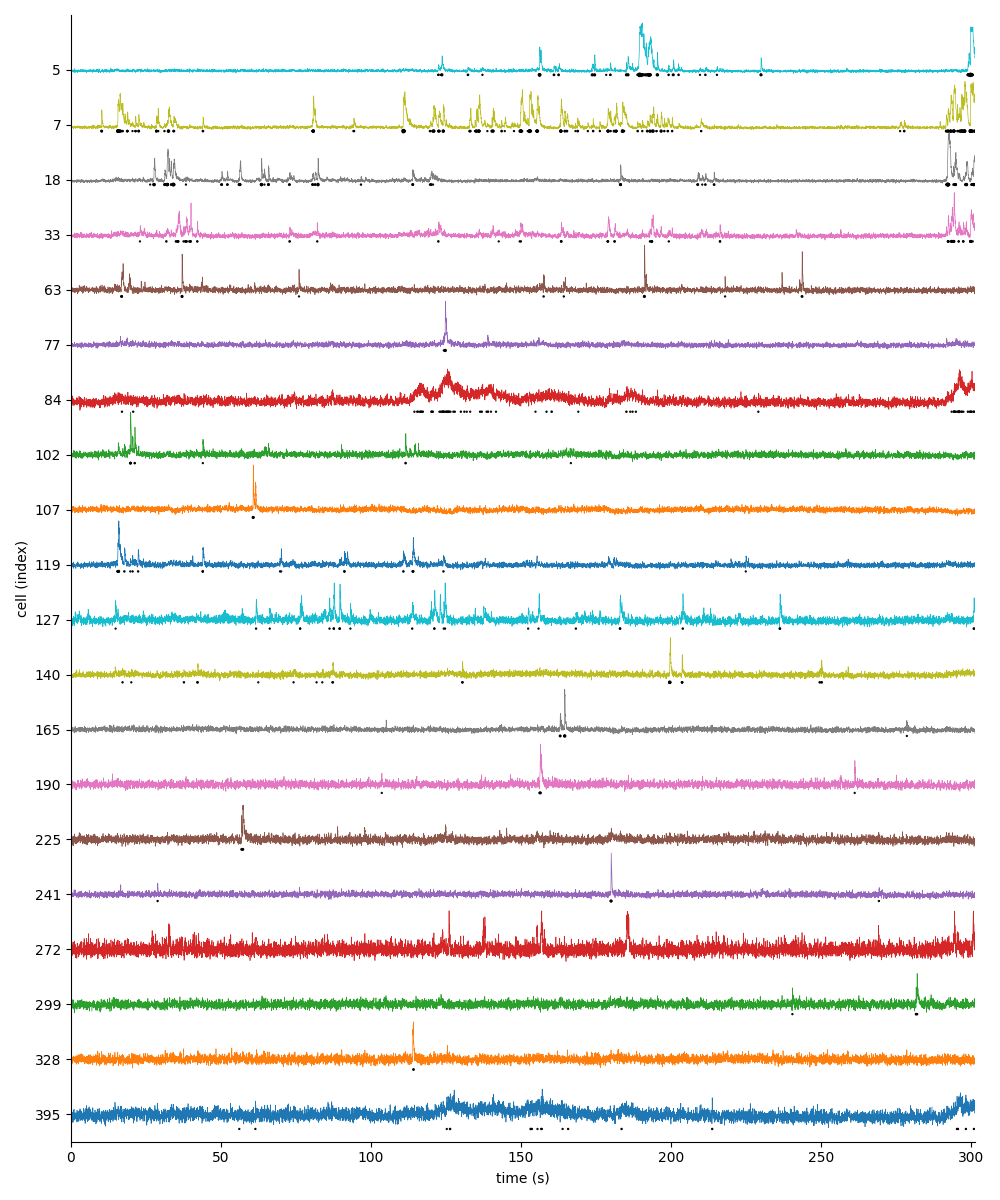 |
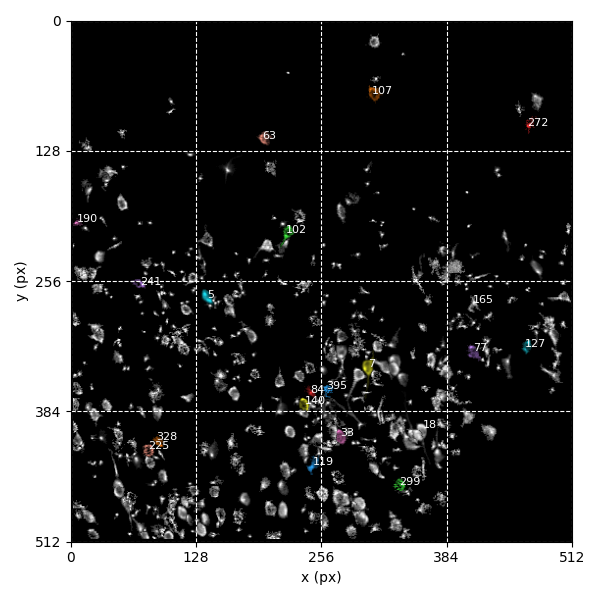 |
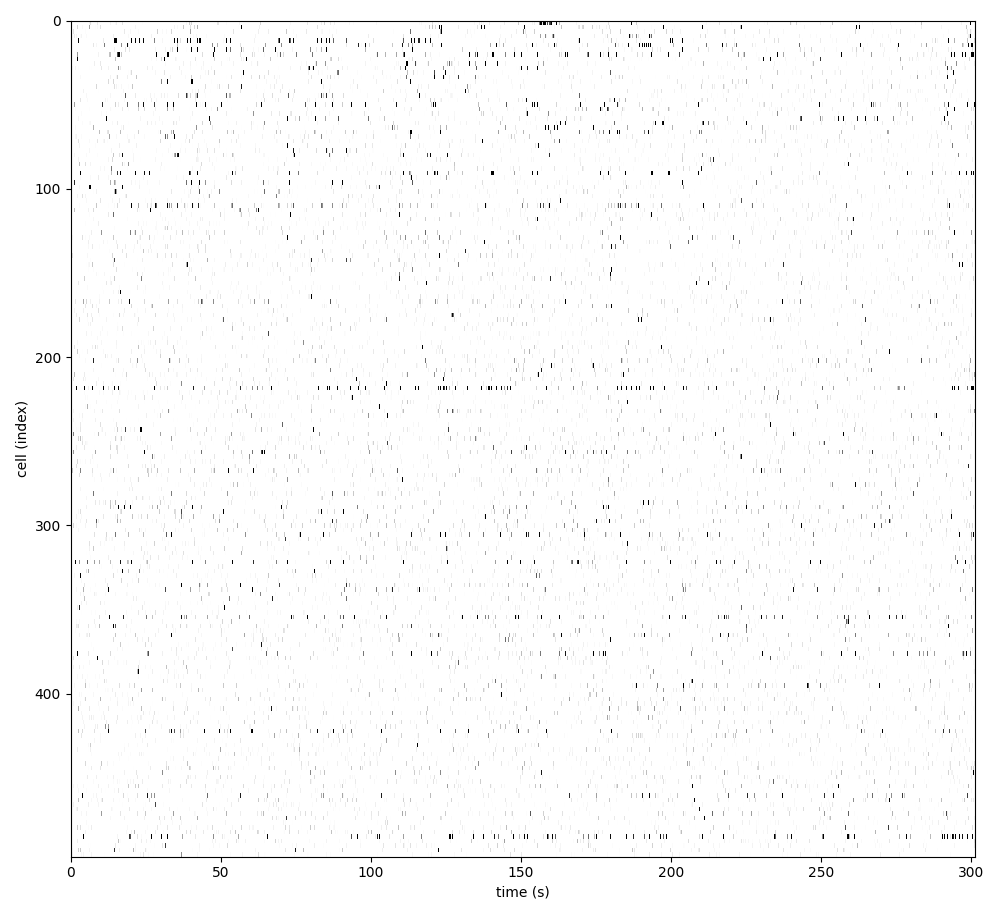 |
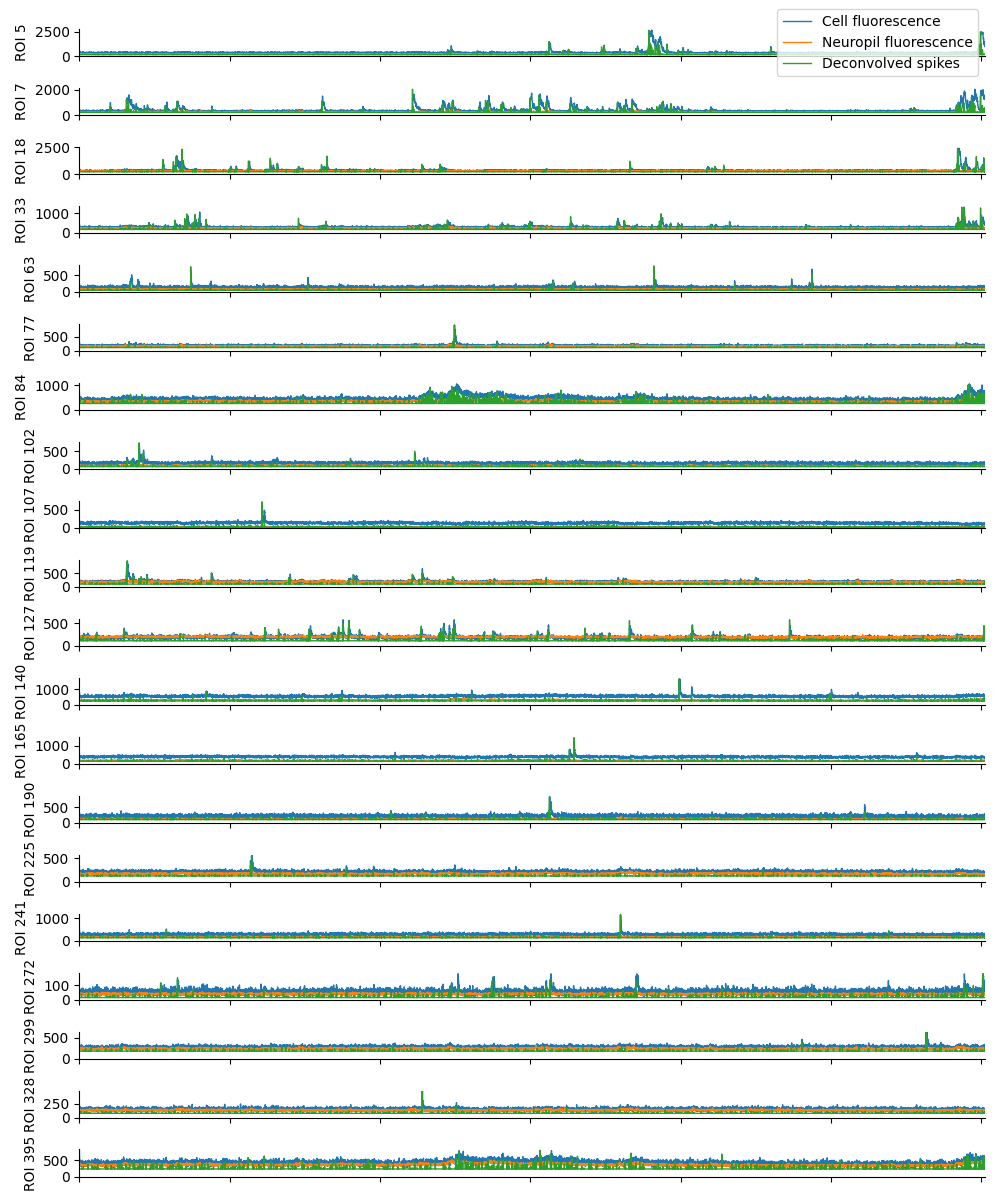 |
For Inscopix .isxd outputs, you can explore 2 preview figures for cellset_raw.isxd:
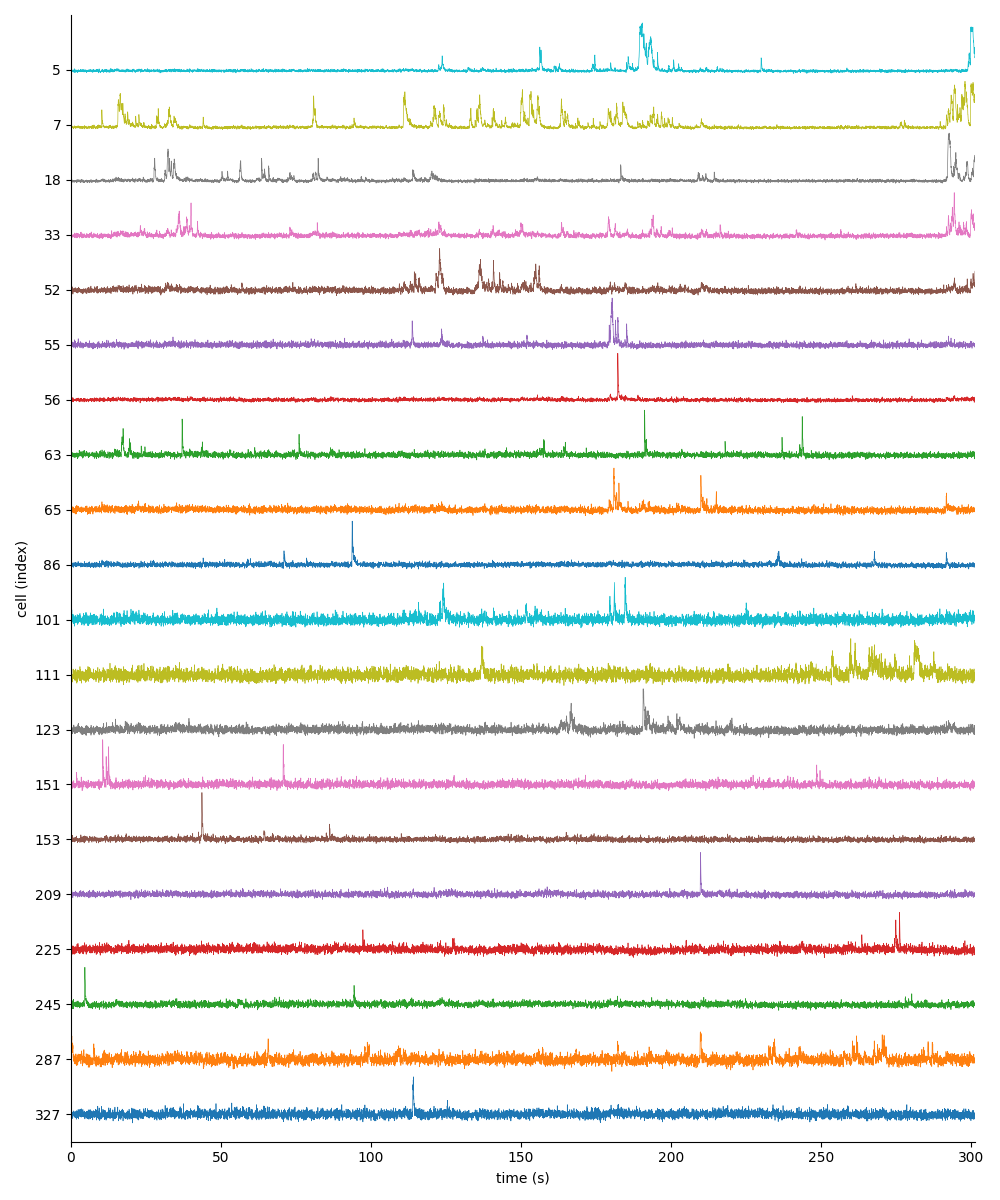 |
 |
and 2 for eventset.isxd (also obtained from processing the same 5-minute 2P movie as above):
 |
 |