CaImAn Cell Extraction Workflow¶
This tool uses 2.0 compute credits per hour.
Overview¶
CaImAn1 is an open-source computational toolbox for large scale Calcium Imaging Analysis, including movie handling, motion correction, source extraction, spike deconvolution and result visualization. The CaImAn cell extraction workflow offered on IDEAS is compatible with both 1P and 2P data. The workflow performs motion correction using the NorMCorre algorithm2, cell identification using the CNMF/CNMF-E algorithm3, and automated component evaluation.
Inputs¶
The workflow supports individual movies and movie series. When a movie series is provided, the movies are concatenated temporally and analyzed as a single movie. When a movie series is processed, the number of output files will match the number of input files. For instance, for each input movie file, a corresponding output cell set file will be generated. While all output cell set files will contain the same spatial footprints, the temporal activity stored in each cell set file will match the timeline of the corresponding input movie.
Once the input movie is selected, an optional parameter file may be selected and used to override the parameters specified in the analysis table. The input files and processing parameters are listed in the table below.
Parameters¶
| Parameter | Required? | Default | Description |
|---|---|---|---|
| Movie(s) | True | N/A | Single movie file or movie series (isxd, tif, tiff, avi) |
| Optional Parameters File | False | N/A | JSON file containing CaImAn processing parameters |
| Overwrite Analysis Table Parameters | True | False | If True and a parameters file has been selected, the analysis table columns will be overwritten by the values contained in the parameters file. |
| fr | False | auto | Imaging rate in frames per second. If set to 'auto', the frame rate will be automatically determined based on file metadata. If no such metadata is available, CaImAn will default to 30 frames per second. |
| decay_time | False | 0.4 | Length of a typical calcium transient in seconds |
| dxy | False | 1.0 | Spatial resolution of the field of view in pixels per um |
| motion_correct | False | True | If True, motion correction will be performed |
| pw_rigid | False | False | If True, piecewise-rigid motion correction will be performed. If False, rigid motion correction will be performed. |
| gSig_filt | False | 3 | Size of the kernel used for high-pass spatial filtering of the data. If unspecified, no spatial filtering will be performed during motion correction. Applying a high-pass spatial filter may improve performance on 1P data by delineating edges and sharpening the images. Such filtering may not be needed for 2P data and could be left unspecified. |
| max_shifts | False | 5 | Maximum shift per dimension in pixels during motion correction |
| strides | False | 48 | The stride controls the shift in pixels between patches of the field of view during piecewise-rigid registration |
| overlaps | False | 24 | Overlap between patches in pixels during piecewise-rigid motion correction |
| max_deviation_rigid | False | 3 | Maximum deviation in pixels between rigid shifts and shifts of individual patches |
| border_nan | False | copy | Strategy for handling NaN values along the boundaries. 'True' will allow NaN values along the boundaries, 'False' will replace any NaN with 0, 'copy' will replace any NaN with the value of the nearest data point, and 'min' will replace any NaN with the minimum value in the image. |
| p | False | 1 | Order of the autoregressive model used to deconvolve the indicator temporal dynamics |
| K | False | auto | Number of components to be found per patch. If set to 'auto', the number of components will automatically be estimated. If 'rf' is unspecified (i.e. single patch), 'K' will correspond to the number of cells in the entire field of view. |
| gSig | False | 3 | Radius of an average neuron in pixels |
| gSiz | False | 7 | Half-size of the bounding box for each neuron |
| merge_thr | False | 0.7 | Trace correlation threshold for merging two components |
| rf | False | 40 | Half-size of a patch in pixels. If unspecified, the entire field of view will be processed as a single patch. |
| stride | False | 20 | Overlap between neighboring patches in pixels |
| tsub | False | 2 | Temporal downsampling factor |
| ssub | False | 1 | Spatial downsampling factor |
| method_init | False | corr_pnr | Method used for initializing cellular components. Use 'corr_pnr' for 1P data, 'greedy_roi' for 2P data, and 'sparse_NMF' for dendritic data. |
| min_corr | False | 0.85 | Minimum value of the correlation image for identifying a candidate component during 'corr_pnr' initialization |
| min_pnr | False | 12 | Minimum value of the peak-to-noise-ratio (pnr) image for identifying a candidate component during 'corr_pnr' initialization |
| ssub_B | False | 2 | Spatial downsampling factor to use when estimating the background activity during 'corr_pnr' initialization |
| ring_size_factor | False | 1.4 | Multiple of the radius of an average neuron (gSig) to use for computing the radius of the ring model used for estimating the background activity during 'corr_pnr' initialization |
| method_deconvolution | False | oasis | Method for solving the constrained deconvolution of temporal traces |
| update_background_components | False | True | If True, the spatial background components will be updated in each iteration of the algorithm |
| del_duplicates | False | True | If True, duplicate components in the overlapping regions between neighboring patches will be deleted. If False, a merging procedure will be used to determine whether the components are similar enough to be merged. |
| nb | False | 0 | Number of global background components. Set to 0 for 1P data. A default value of 1 may be used for 2P data. |
| low_rank_background | False | False | If True, the background will be updated using low-rank approximation. If False, all nonzero elements of the background components will be updated using the Hierarchical Alternating Least Squares (HALS) algorithm. Set to False for 1P data and True for 2P data. |
| nb_patch | False | 0 | Number of local background components per patch. Set to 0 for 1P data. A default value of 1 may be used for 2P data. |
| rolling_sum | False | True | If True, a rolling sum will be used for determining candidate centroids during 'greedy_roi' initialization. If False, a full sum will be used. |
| only_init | False | True | If True, the values after initialization will be returned |
| normalize_init | False | False | If True, pixel values will be normalized by their respective median intensity across the input movies |
| center_psf | False | True | If True, merge overlapping components by taking the best neuron. Set to True for 1P data and False for 2P data. |
| bas_nonneg | False | False | If True, a non-negative baseline will be used. If False, the baseline will be greater than or equal to the minimum value, which could be negative. |
| border_pix | False | 0 | Number of pixels to exclude around each border |
| refit | False | False | If True, the initial estimates will be refined by re-running the CNMF algorithm seeded on the spatial estimates from the previous step |
| SNR_lowest | False | 0.5 | Minimum signal-to-noise ratio required for a trace to be accepted. Traces with a signal-to-noise ratio below this will be rejected |
| min_SNR | False | 3 | Trace signal-to-noise ratio threshold. Traces with signal-to-noise ratios above this will be accepted |
| rval_lowest | False | -1 | Minimum spatial correlation required for a component to be accepted. Components with spatial correlation below this will be rejected |
| rval_thr | False | 0.85 | Spatial correlation threshold. Components with spatial correlation higher than this will be accepted |
| use_cnn | True | False | If True, a pretrained convolutional neural network (CNN) will be used for accepting/rejecting components. The model was trained on 2P data and may not be applicable to 1P data. |
| cnn_lowest | False | 0.1 | Minimum CNN classifier score required to accept components. Components with scores lower than this will be rejected |
| gSig_range | False | N/A | List of neuron sizes to compare when using the CNN classifier. If unspecified, the CNN classifier will be applied using a neuron size equal to 'gSig'. If multiple values are provided, the CNN classifier will be tested will each one and the predictions of the best-performing run returned. |
| min_cnn_thr | False | 0.9 | CNN classifier score threshold. Components with scores higher than this will be accepted |
| n_processes | False | 7 | Number of processes used to analyze patches in parallel |
Optional Parameters File Format¶
A json file may be uploaded to IDEAS and used to specify CaImAn's processing parameters.
To override the analysis table parameters with the parameters specified in the json file supplied,
the Overwrite Analysis Table Parameters parameter must be set to True.
Sample parameter files from CaImAn are included below for reference.
{
"data": {
"fr": 10,
"decay_time": 0.4
},
"init": {
"method_init": "corr_pnr",
"K": null,
"gSig": [3, 3],
"gSiz": [13, 13],
"ssub": 1,
"tsub": 2,
"nb": 0,
"min_corr": 0.8,
"min_pnr": 10,
"normalize_init": false,
"center_psf": true,
"ssub_B": 2,
"ring_size_factor": 1.4
},
"motion": {
"pw_rigid": false,
"max_shifts": [5, 5],
"gSig_filt": [3, 3],
"strides": [48, 48],
"overlaps": [24, 24],
"max_deviation_rigid": 3,
"border_nan": "copy"
},
"preprocess": {
"p": 1
},
"temporal": {
"p": 1,
"method_deconvolution": "oasis"
},
"spatial": {
"update_background_components": true
},
"patch": {
"rf": 40,
"stride": 20,
"only_init": true,
"nb_patch": 0,
"low_rank_background": null,
"del_duplicates": true
},
"merging": {
"merge_thr": 0.7
},
"quality": {
"min_SNR": 2.5,
"rval_thr": 0.85,
"use_cnn": false
}
}
{
"data": {
"fr": 30,
"decay_time": 0.4,
"dxy": [2.0, 2.0]
},
"init": {
"nb": 2,
"K": 4,
"gSig": [4, 4],
"method_init": "greedy_roi",
"rolling_sum": true,
"ssub": 2,
"tsub": 2
},
"motion": {
"pw_rigid": true,
"max_shifts": [6, 6],
"strides": [50, 50],
"overlaps": [24, 24],
"max_deviation_rigid": 3,
"border_nan": "copy"
},
"preprocess": {
"p": 1
},
"temporal": {
"p": 1
},
"patch": {
"rf": 15,
"stride": 6,
"only_init": true
},
"merging": {
"merge_thr": 0.85
},
"quality": {
"min_SNR": 2,
"rval_thr": 0.85,
"use_cnn": true,
"min_cnn_thr": 0.99,
"cnn_lowest": 0.1
}
}
1P vs 2P Data Processing¶
The workflow is compatible with both 1P and 2P data.
The parameters that control the initialization step as well as how to model background fluctuations
can be adjusted based on the input data.
The default parameters are set for processing 1P data.
The table below summarizes the parameters that may be adjusted based on the desired workflow.
None means the parameter is unspecified and the corresponding analysis table cell is left empty.
Note that K must be set based on the expected number of components per patch for 2P data.
| Parameter | 1P | 2P |
|---|---|---|
| gSig_filt | 3 | None |
| K | auto | 4 |
| method_init | corr_pnr | greedy_roi |
| del_duplicates | true | false |
| nb | 0 | 2 |
| low_rank_background | false | true |
| nb_patch | 0 | 1 |
| normalize_init | false | true |
| center_psf | true | false |
| bas_nonneg | false | true |
| use_cnn | false | true |
Outputs¶
The workflow will produce the 4 outputs listed below.
CaImAn Output¶
hdf5 file containing the model and data produced by CaImAn.
This includes the model itself and the results of the algorithm, including cellular footprints,
temporal dynamics, spikes, noise estimates, background activity, residuals, and more.
The data preview includes the initialization images used by the algorithm as well as the spatial footprints and raw temporal traces of putative cells detected by CaImAn.
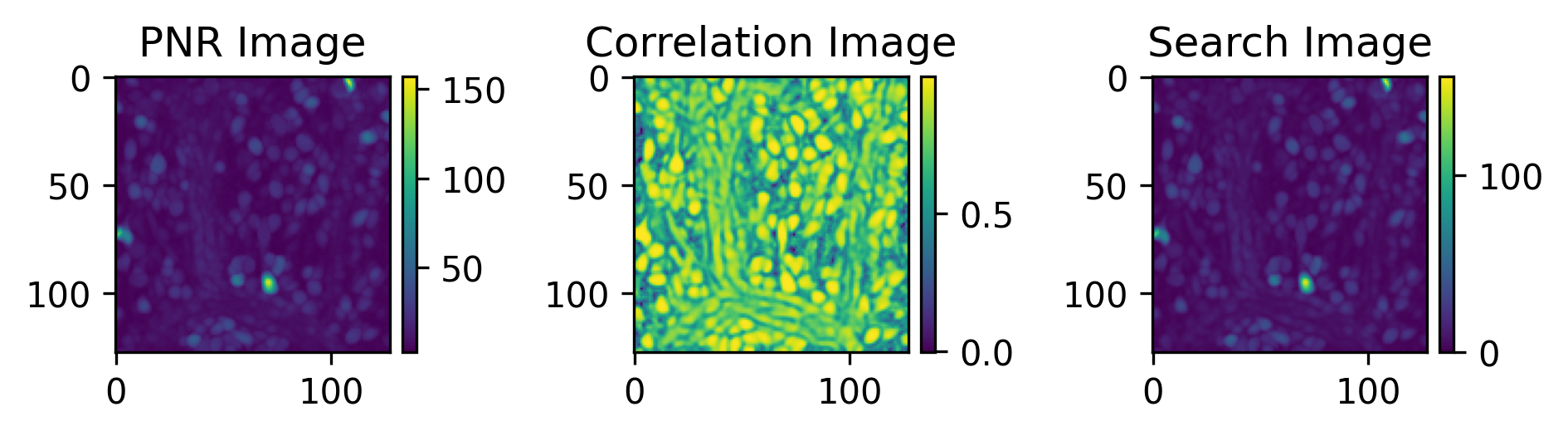
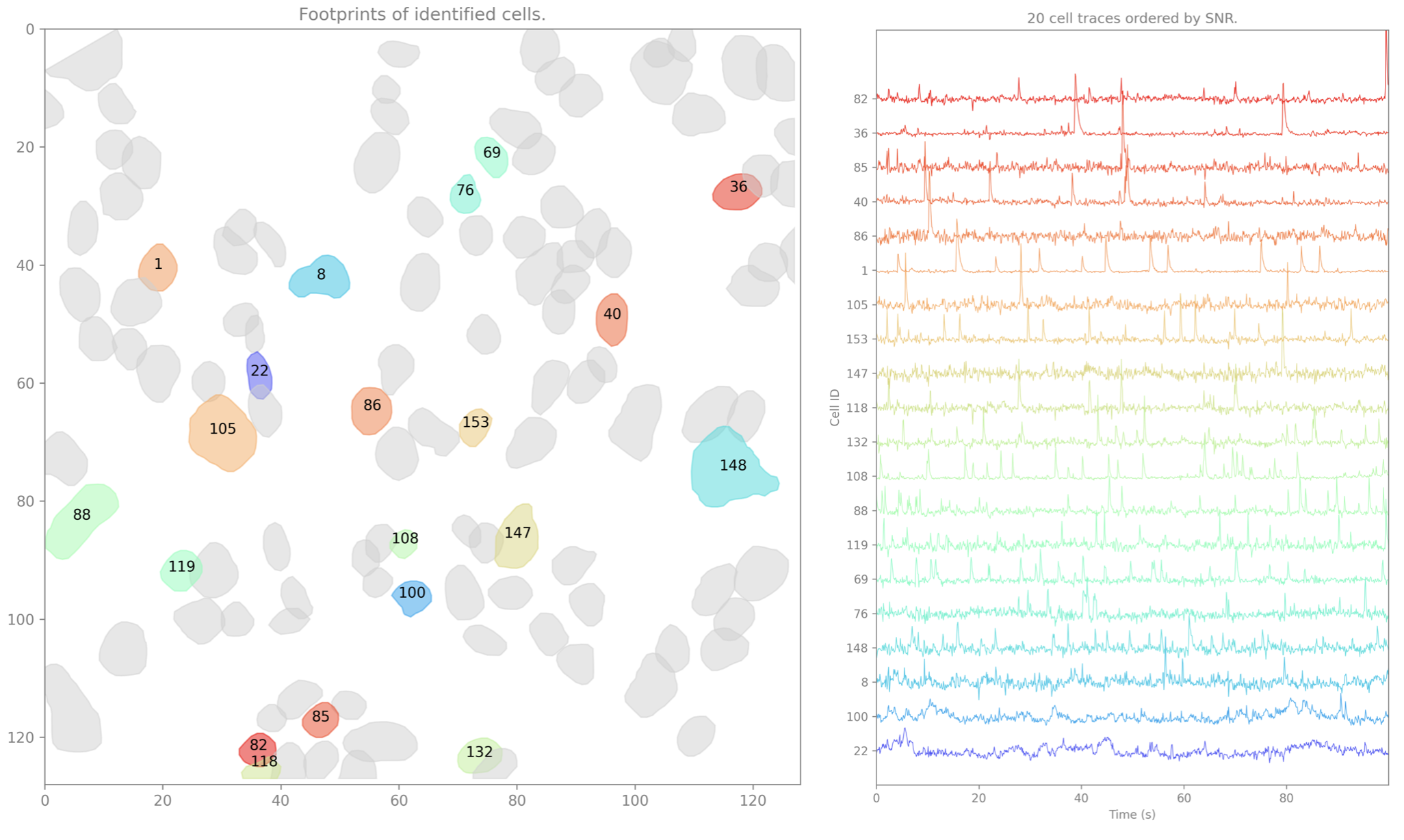
Raw Cell Set(s)¶
isxd cell set(s) containing the spatial footprints and raw temporal traces of the cells identified by CaImAn.

Denoised Cell Set(s)¶
isxd cell set(s) containing the spatial footprints and denoised temporal traces of the cells identified by CaImAn.
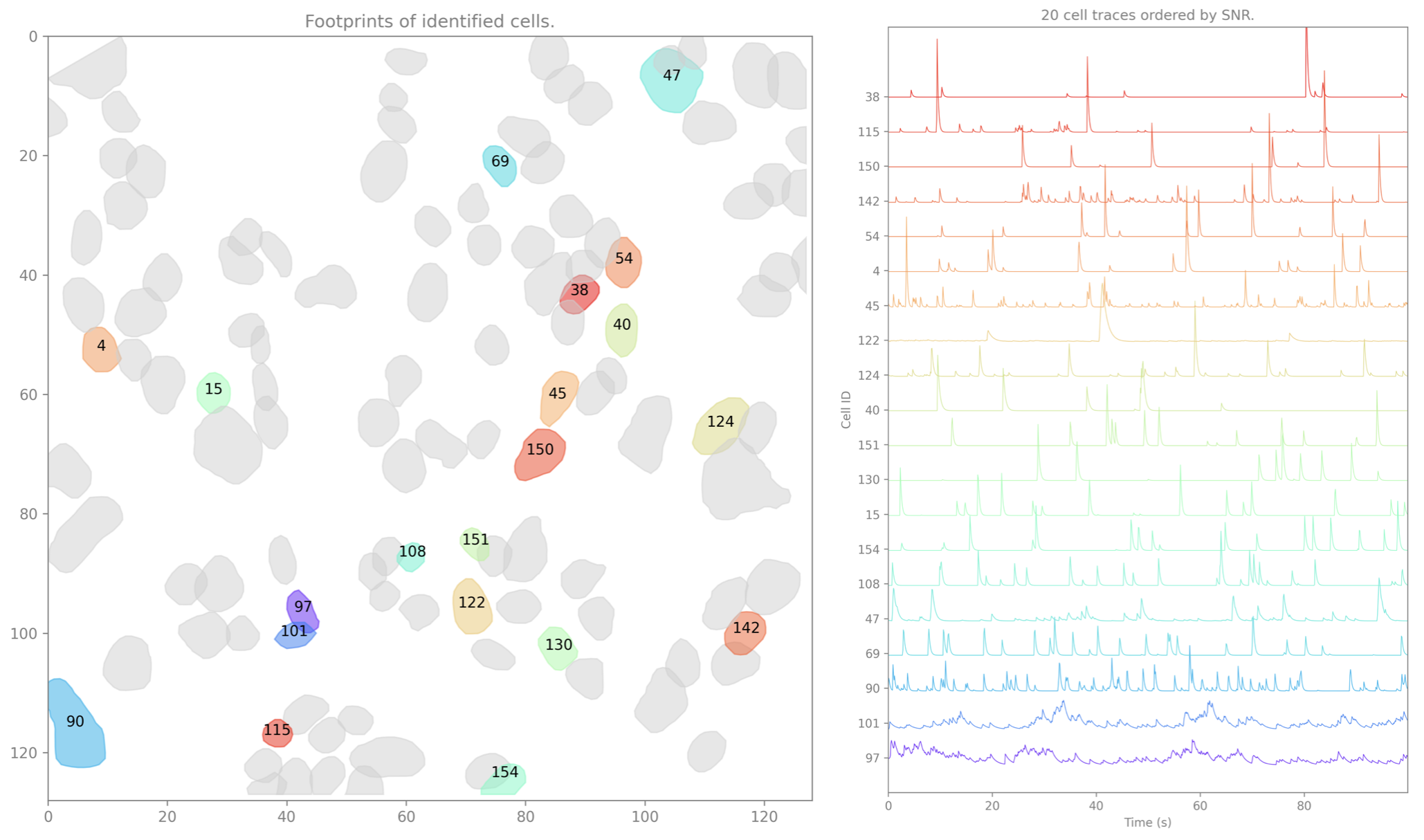
Spike Event Set(s)¶
isxd event set(s) containing the neural events identified by CaImAn.
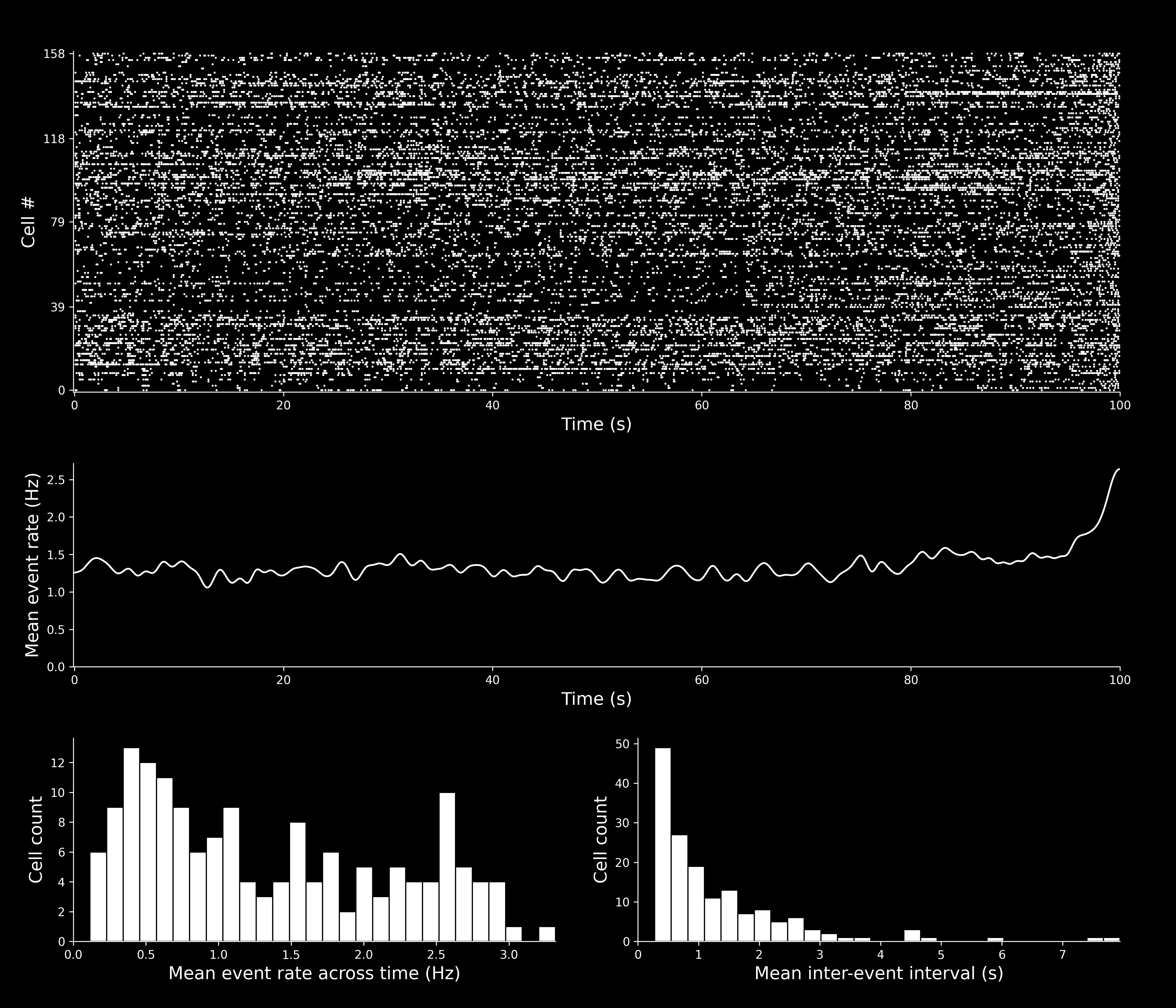
-
Giovannucci 2018, CaImAn an open source tool for scalable calcium imaging data analysis ↩
-
Pnevmatikakis 2017, NoRMCorre: An online algorithm for piecewise rigid motion correction of calcium imaging data ↩
-
Zhou 2018, Efficient and accurate extraction of in vivo calcium signals from microendoscopic video data ↩