suite2p ROI classification¶
This tool uses 0.5 compute credits per hour.
Overview¶
This suite2p ROI classification tool is a wrapper for the suite2p function classification.classify().
You can find more information on this processing step in the suite2p docs.
suite2p ROI classification curates extracted ROIs, potentially belonging to different cell compartments (cell body aka soma, dendrites, and axon), using either a built-in classifier or a user-provided one, to only keep cell bodies (aka somata) for further analyses.
Note that this tool constitutes the fifth step of Run suite2p pipeline.
Input files¶
| Source Parameter | File Type | File Format |
|---|---|---|
| Extraction Statistics File | suite2p_data | npy |
| Parameters File | config | npy |
| Optional Classifier File | suite2p_classifier | npy |
Parameters¶
| Parameter | suite2p name | Required? | Default | Description |
|---|---|---|---|---|
| Extraction Statistics File | - | True | N/A | Input cell statistics file, as outputted by the ROI extraction step [ |
| Parameters File | - | True | N/A | Input suite2p parameters file, as outputted by the ROI extraction tool [ |
| Optional Classifier File | classifier_path | False | N/A | [from suite2p docs] "Path to classifier file you want to use for cell classification." [.npy] |
| Soma Crop | soma_crop | False | True | [from suite2p docs] "Specifies whether to crop dendrites for cell classification stats (e.g., compactness)." [bool] |
| FOV vmin Percentile | - | False | 0 | Minimum value for the colormap range, as percentile of the FOV fluorescence [float, [0 |
| FOV vmax Percentile | - | False | 99 | Maximum value for the colormap range, as percentile of the FOV fluorescence [float, ( |
| FOV Colormap | - | False | plasma | Colormap for plotting the FOV [str] |
| Show Grid on FOV | - | False | True | Whether or not to show the grid on FOVs [bool] |
| FOV Ticks Step | - | False | 128 | Step for the x- and y-ticks [int, >0] |
Output files¶
| File name | File type | Notes |
|---|---|---|
| iscell.npy | NumPy file | Contains classification labels for all extracted ROIs, as a 1D array. |
| ops_ROI_classification.npy | NumPy file | Contains a dictionary of parameters populated by suite2p ROI classification. |
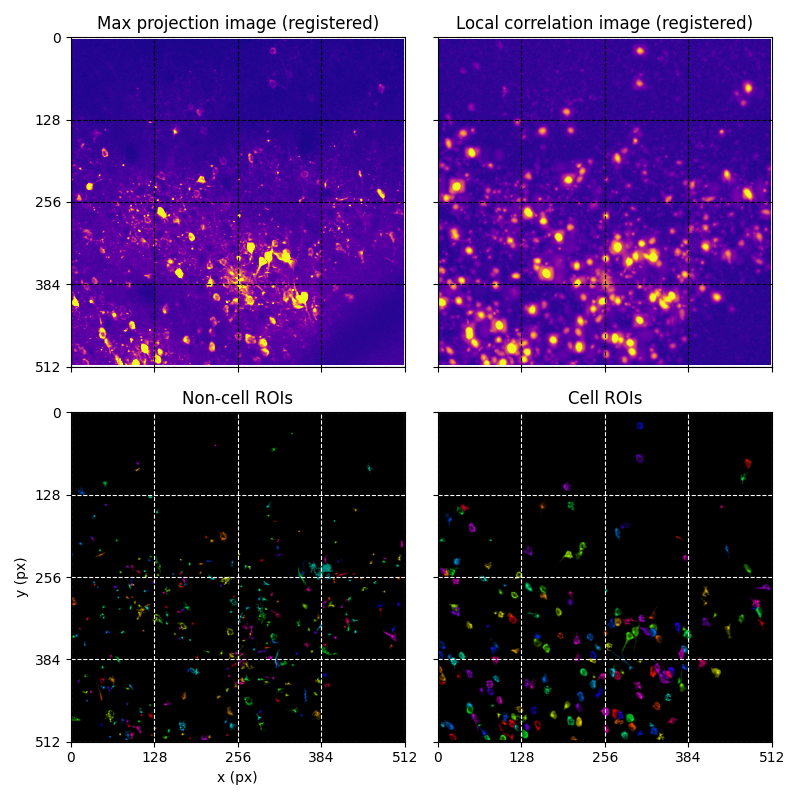
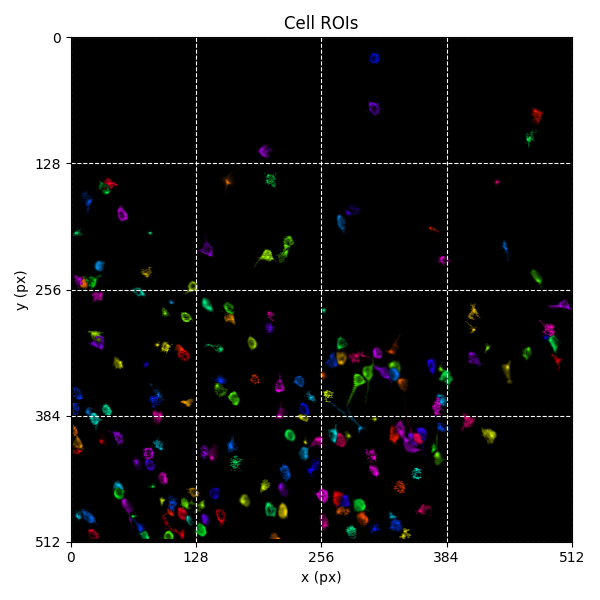
You can explore 2 preview figures for iscell.npy.
You can find below an example of these previews obtained from processing a 5-minute 2P movie of mouse cortex (data courtesy of Dr. Ahmet Arac, MD, at UCLA):
 |
 |