suite2p spike deconvolution¶
This tool uses 0.5 compute credits per hour.
Overview¶
This suite2p spike deconvolution tool is a wrapper for the suite2p functions extraction.preprocess() and extraction.oasis(), called in run_s2p().
You can find more information on this processing step in the suite2p docs.
suite2p spike deconvolution infers underlying spike events from the neuropil-corrected fluorescence traces using spike deconvolution based on the OASIS algorithm.
Note that this tool constitutes the first step of Run suite2p pipeline.
Input files¶
| Source Parameter | File Type | File Format |
|---|---|---|
| Fluorescence Traces File | suite2p_data | npy |
| Neuropil Fluorescence Traces File | suite2p_data | npy |
| Parameters File | config | npy |
Parameters¶
| Parameter | suite2p name | Required? | Default | Description |
|---|---|---|---|---|
| Fluorescence Traces File | - | True | N/A | Input fluorescence traces file, as outputted by the ROI extraction step [ |
| Neuropil Fluorescence Traces File | - | True | N/A | Input neuropil fluorescence traces file, as outputted by the ROI extraction step [ |
| Parameters File | - | True | N/A | Input suite2p parameters file, as outputted by the ROI extraction tool [ |
| Tau | tau | False | 1.0 | [from suite2p docs] "The timescale of the sensor (in seconds), used for deconvolution kernel. The kernel is fixed to have this decay and is not fit to the data. We recommend: 0.7 for GCaMP6f; 1.0 for GCaMP6m; 1.25-1.5 for GCaMP6s." [float, =0.001] |
| Neuropil Coefficient | neucoeff | False | 0.7 | [from suite2p docs] "Neuropil coefficient for all ROIs." [float, =0] |
| Baseline | baseline | False | maximin | [from suite2p docs] "How to compute the baseline of each trace. This baseline is then subtracted from each cell. 'maximin' computes a moving baseline by filtering the data with a Gaussian of width ops['sig_baseline'] * ops['fs'], and then minimum filtering with a window of ops['win_baseline'] * ops['fs'], and then maximum filtering with the same window. 'constant' computes a constant baseline by filtering with a Gaussian of width ops['sig_baseline'] * ops['fs'] and then taking the minimum value of this filtered trace. 'constant_percentile' computes a constant baseline by taking the ops['prctile_baseline'] percentile of the trace." [str, ['maximin', 'constant', 'constant_percentile']] |
| Window Baseline | win_baseline | False | 60.0 | [from suite2p docs] "Window for maximin filter in seconds." [float, 0] |
| Sig Baseline | sig_baseline | False | 10.0 | [from suite2p docs] "Gaussian filter width in seconds, used before maximin filtering or taking the minimum value of the trace, ops['baseline'] = 'maximin' or 'constant'." [float, 0] |
| Percentile Baseline | prctile_baseline | False | 8.0 | [from suite2p docs] "Percentile of trace to use as baseline if ops['baseline'] = 'constant_percentile'." [float, 0] |
| Number of Sample Cells | - | False | 20 | Number of sample cells for the spike deconvolution preview [int, >0] |
| Random Seed | - | False | 0 | Random seed for selecting sample cells [int, >=0] |
| Show Non-Sample Cells' Footprints | - | False | True | Whether or not to show footprints of non-sample cells on the cell footprint FOV image. If False, only footprints of sample cells are displayed [bool] |
Output files¶
| File name | File type | Notes |
|---|---|---|
| spks.npy | NumPy file | Contains deconvolved spikes for all extracted ROIs, as a 2D array. |
| ops_spike_deconvolution.npy | NumPy file | Contains a dictionary of parameters populated by suite2p spike deconvolution. |
You can explore 3 preview figures for spks.npy.
You can find below an example of these previews obtained from processing a 5-minute 2P movie of mouse cortex (data courtesy of Dr. Ahmet Arac, MD, at UCLA):
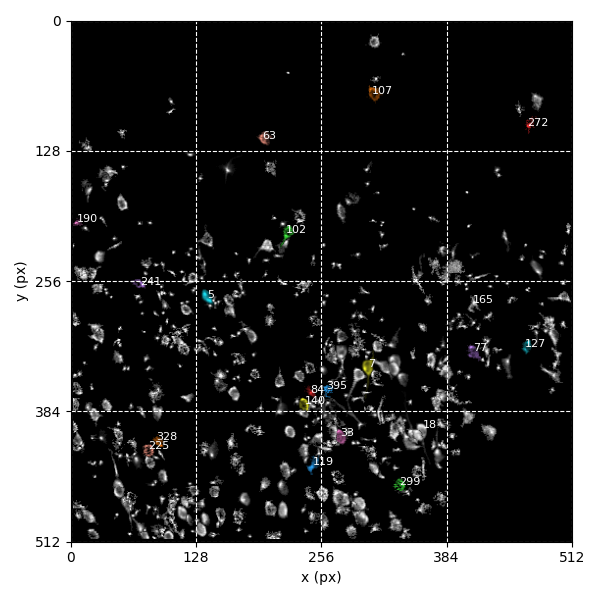 |
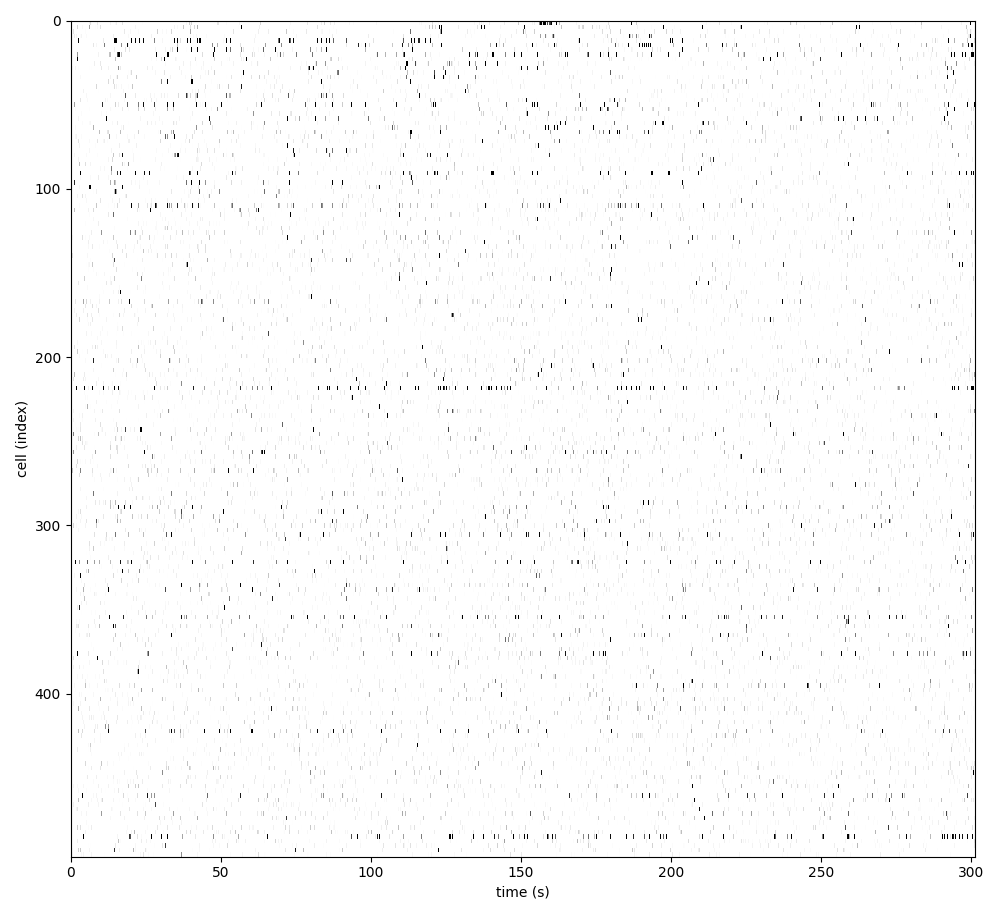 |
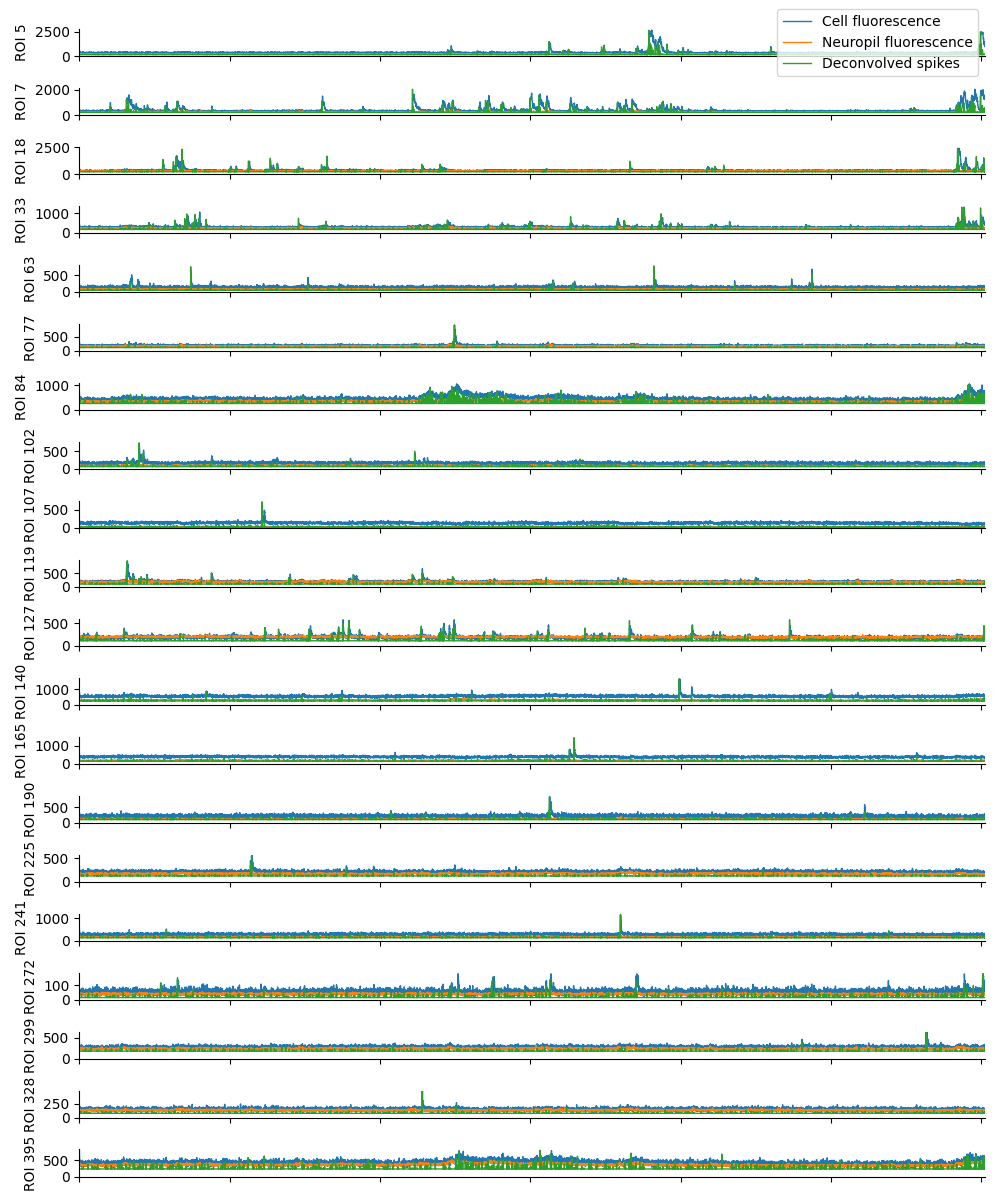 |