suite2p ROI detection¶
This tool uses 1.0 compute credits per hour.
Overview¶
This suite2p ROI detection tool is a wrapper for the suite2p function detection.detection_wrapper().
You can find more information on this processing step in the suite2p docs.
suite2p ROI detection detects Regions-Of-Interest (ROIs) using computationally efficient, semi-constrained matrix factorization.
Note that this tool constitutes the third step of Run suite2p pipeline.
Input files¶
| Source Parameter | File Type | File Format |
|---|---|---|
| Registered Binary Movie | suite2p_data | bin |
| Parameters File | config | npy |
| Optional Classifier File | suite2p_classifier | npy |
Parameters¶
| Parameter | suite2p name | Required? | Default | Description |
|---|---|---|---|---|
| Registered Binary Movie | - | True | N/A | Input registered suite2p binary movie [.bin] |
| Parameters File | - | True | N/A | Input suite2p parameters file, as outputted by the registration tool [ |
| Optional Classifier File | classifier_path | False | N/A | [from suite2p docs] "Path to classifier file you want to use for cell classification." [.npy] |
| Tau | tau | False | 1.0 | [from suite2p docs] "The timescale of the sensor (in seconds), used for deconvolution kernel. The kernel is fixed to have this decay and is not fit to the data. We recommend: 0.7 for GCaMP6f; 1.0 for GCaMP6m; 1.25-1.5 for GCaMP6s." [float, =0.001] |
| Sparse Mode | sparse_mode | False | True | [from suite2p docs] "Whether or not to use sparse_mode cell detection." [bool] |
| Spatial Scale | spatial_scale | False | 0 | [from suite2p docs] "What the optimal scale of the recording is in pixels. if set to 0, then the algorithm determines it automatically (recommend this on the first try). If it seems off, set it yourself to the following values: 1 (=6 pixels), 2 (=12 pixels), 3 (=24 pixels), or 4 (=48 pixels)." [int, [0 4]] |
| Connected | connected | False | True | [from suite2p docs] "Whether or not to require ROIs to be fully connected (set to 0 for dendrites/boutons)." [bool] |
| Threshold Scaling | threshold_scaling | False | 1.0 | [from suite2p docs] "This controls the threshold at which to detect ROIs (how much the ROIs have to stand out from the noise to be detected). if you set this higher, then fewer ROIs will be detected, and if you set it lower, more ROIs will be detected." [float] |
| Spatial HP Detect | spatial_hp_detect | False | 25 | [from suite2p docs] "Window for spatial high-pass filtering for neuropil subtracation before ROI detection takes place." [int, 0] |
| Max Overlap | max_overlap | False | 0.75 | [from suite2p docs] "We allow overlapping ROIs during cell detection. After detection, ROIs with more than ops['max_overlap'] fraction of their pixels overlapping with other ROIs will be discarded. Therefore, to throw out NO ROIs, set this to 1.0." [float, [0 1]] |
| High Pass | high_pass | False | 100 | [from suite2p docs] "Running mean subtraction across time with window of size 'high_pass'. Values of less than 10 are recommended for 1P data where there are often large full-field changes in brightness." [int, 0] |
| Smooth Masks | smooth_masks | False | True | [from suite2p docs] "Whether to smooth masks in final pass of cell detection. This is useful especially if you are in a high noise regime." [bool] |
| Max Iterations | max_iterations | False | 20 | [from suite2p docs] "How many iterations over which to extract cells - at most ops['max_iterations'], but usually stops before due to ops['threshold_scaling'] criterion." [int, 0] |
| Number of Binned Frames | nbinned | False | 5000 | [from suite2p docs] "Maximum number of binned frames to use for ROI detection." [int, 0] |
| Denoise | denoise | False | False | [from suite2p docs] "Whether or not binned movie should be denoised before cell detection in sparse_mode. If True, make sure to set ops['sparse_mode'] is also set to True." [bool] |
| Anatomical Only | anatomical_only | False | 0 | [from suite2p docs] "If greater than 0, specifies what to use Cellpose on. 1: Will find masks on max projection image divided by mean image. 2: Will find masks on mean image 3: Will find masks on enhanced mean image 4: Will find masks on maximum projection image." [int, [0 4]] |
| Diameter | diameter | False | 0 | [from suite2p docs] "Diameter that will be used for cellpose. If set to zero, diameter is estimated." [int, =0] |
| Cellprob Threshold | cellprob_threshold | False | 0 | [from suite2p docs] "Specifies threshold for cell detection that will be used by cellpose." [float] |
| Flow Threshold | flow_threshold | False | 1.5 | [from suite2p docs] "Specifies flow threshold that will be used for cellpose." [float] |
| Spatial HP CP | spatial_hp_cp | False | 0 | [from suite2p docs] "Window for spatial high-pass filtering of image to be used for cellpose." [int, =0] |
| Pretrained Model | pretrained_model | False | cyto | [from suite2p docs] "Path to pretrained model or string for model type (can be user's model )." See cellpose docs for a list of available models, e.g., 'cyto', 'cyto2', 'cyto3', 'nuclei'. [str] |
| Preclassify | preclassify | False | 0.0 | [from suite2p docs] "Apply classifier before signal extraction with probability threshold of 'preclassify'. If this is set to 0.0, then all detected ROIs are kept and signals are computed." [float, =0] |
| Channel 2 Threshold | chan2_thres | False | 0.65 | [from suite2p docs] "Threshold for calling an ROI "detected" on a second channel." [float, =0] |
| FOV vmin Percentile | - | False | 0 | Minimum value for the colormap range, as percentile of the FOV fluorescence [float, [0 |
| FOV vmax Percentile | - | False | 99 | Maximum value for the colormap range, as percentile of the FOV fluorescence [float, ( |
| FOV Colormap | - | False | plasma | Colormap for plotting the FOV [str] |
| Show Grid on FOV | - | False | True | Whether or not to show the grid on FOVs [bool] |
| FOV Ticks Step | - | False | 128 | Step for the x- and y-ticks [int, >0] |
Output files¶
| File name | File type | Notes |
|---|---|---|
| stat_ROI_detection.npy | NumPy file | Contains a dictionary of spatial and functional statistics for the detected ROIs. |
| ops_ROI_detection.npy | NumPy file | Contains a dictionary of parameters populated by suite2p ROI detection. |
You can explore 2 preview figures for stat_ROI_detection.npy.
You can find below an example of these previews obtained from processing a 5-minute 2P movie of mouse cortex (data courtesy of Dr. Ahmet Arac, MD, at UCLA):
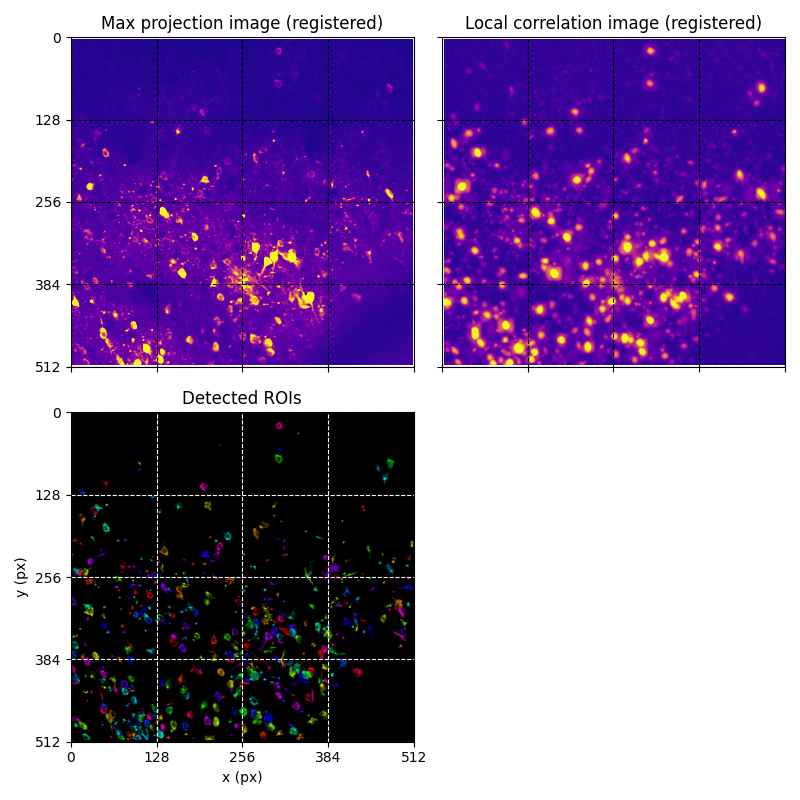 |
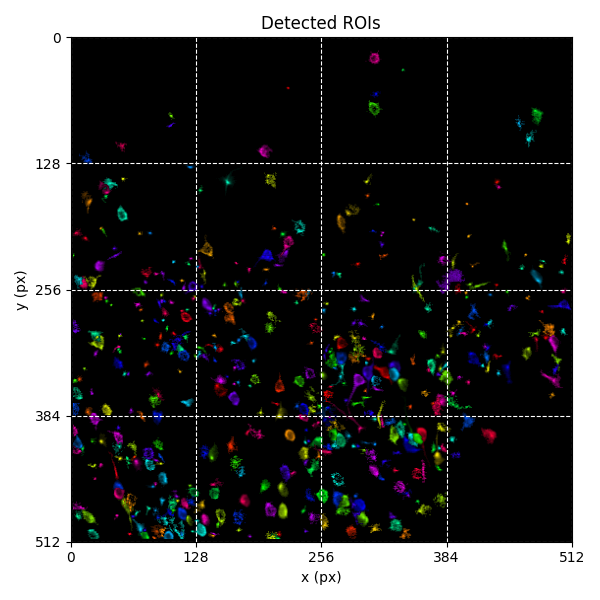 |