Peri-Event Analysis Workflow¶
This tool uses 1.0 compute credits per hour.
Overview¶
The peri-event analysis workflow quantifies how neural signals vary before and after event timestamps. Specifically, this analysis computes the change in neural activity of both the entire population of neurons and each individual neuron, classifying them into up-modulated, down-modulated, or non-modulated sub-populations.
We calculate the peri-event change in activity (Δ activity) by taking the difference between the post-event and pre-event activity, and compare it to a null distribution generated by circularly permuting the event times relative to the neural time series. This comparison is non-parametric and makes few assumptions about the underlying distributions of neural activity or events.
Peri-event analysis is a broadly applicable workflow, and has been extensively used in the field of systems neuroscience for the analysis of signals at the sensory1 2, cognitive3 4 5 6 and motor research7 8 9.
The Compare Peri-Event Activity Across Epochs tool can also be used to quantify and compare peri-event activity across a number of epochs.
Input Files¶
To run the peri-event analysis workflow, users muse have a Cellset and a corresponding Events file.
The Cellsetinput contains time series traces for each cell in the field-of-view of the recording along with the
corresponding footprints.
An Events file contains information about when events occurred. Note that each event in this
context refers to a single time event point.
Event files supported by this tool can be generated using Convert CSV File to Events or Detect Events from GPIO Data.
| Source Parameter | File Type | File Format |
|---|---|---|
| Input cell sets | cell_set | isxd |
| Input events | events, timestamp_events | h5, h5 |
Parameters¶
| Parameter | Required? | Default | Description |
|---|---|---|---|
| Input cell sets | True | N/A | Select cell sets to use for analysis |
| Input events | True | N/A | Select event data to use for analysis |
| Event type | True | N/A | Select type of event to analyze from the column names of the event file |
| Statistical window (pre, start) | True | -1 | Start of time window in seconds before each event to use for statistical tests |
| Statistical window (pre, end) | True | 0 | End of time window in seconds before each event to use for statistical tests |
| Statistical window (post, start) | True | 0 | Start of time window in seconds after each event to use for statistical tests |
| Statistical window (post, end) | True | 1 | End of time window in seconds after each event to use for statistical tests |
| Number of random shuffles | True | 1000 | Number of random shuffles of event times to perform for constructing null distribution |
| Significance threshold | True | 0.05 | p-value threshold for classifying neurons as up- or down-modulated |
| Seed | True | 0 | Seed for the random generator used to shuffle event indices |
| Temporal downsampling factor | True | 1 | Temporal downsampling factor to apply to the activity traces before running the peri-event analysis |
| Visual window (pre) | True | -2 | Time in seconds to visualize before each event in output figures (not used for statistics) |
| Visual window (post) | True | 2 | Time in seconds to visualize after each event in output graphs (not used for statistics) |
| Population activity (y-axis range) | False | auto | y-axis range (z-score) applied to the event-aligned population activity plot specified as 'min,max' (e.g. -1,1) or 'auto' |
| Activity heatmap (colormap range) | False | auto | Colormap range (z-score) applied to the activity heatmap specified as 'min,max' (e.g. -1,1) or 'auto' |
| Activity by modulation group (y-axis range) | False | auto | y-axis range (z-score) applied to the event-aligned activity by modulation plot specified as 'min,max' (e.g. -1,1) or 'auto' |
| Modulation colors | False | green,blue,black | Colors that represent up-, down- and non-modulation respectively |
| Neural activity trace color | False | #1f77b4 | The mean and sem trace color for event-aligned population activity |
| Activity heatmap colormap | False | coolwarm | Colormap applied to the activity heatmap |
Tip
-
To quickly view and navigate between parameters, select the ‘Jump to Parameter’ button at the bottom of the page.
-
Hover the mouse over the column name to get a short description of the parameter.
Algorithm Description¶
The workflow is summarized using a flowchart below.
The workflow processes each event type independently. For each event type, the neural activity is first z-scored (performed individually for each neuron), and then the Δ activity is computed at two levels: population level and single-cell level. The Δ activity of all neurons is plotted as a heat-map (Panel A), and the true population average activity is plotted against the null distribution (Panel B).
At both population and single cell level, modulation is determined by comparing the Δ activity to a null distribution constructed by randomly circularly permuting the event times relative to the neural time series. This process is repeated according to Number of random shuffles. For each cell (and for the population) we compute two single-tailed bootstrap probabilities: the fraction of the null distribution greater than the true Δ (p_high) and the fraction less than the true Δ (p_low). The reported p-value is min(p_high, p_low) in [0, 0.5], with resolution 1 / Number of random shuffles (e.g., 1000 shuffles ⇒ 0.001; perfectly extreme cases yield 0). Modulation calls apply one-tailed thresholds at α/2 to implement a two-tailed decision: up-modulated if p_high < α/2, down-modulated if p_low < α/2, otherwise non-significant.
After each individual cell has been classified, they are grouped into up-modulated, down-modulated, and non-modulated subpopulations. The average activity of each subpopulation is plotted (Panel C). The spatial locations of the modulated cells are also plotted (Panel D).
Cell Statuses¶
Individual cell statuses stored in the input cell set are used to determine which cells to include in the analysis. If there are accepted cells, only accepted cells will be used. If there are no accepted cells, only undecided cells will be used. If all cells are rejected, the workflow will raise an error to inform the user.
Output figures¶
Panel A: Event-Aligned single cell activity heatmap¶
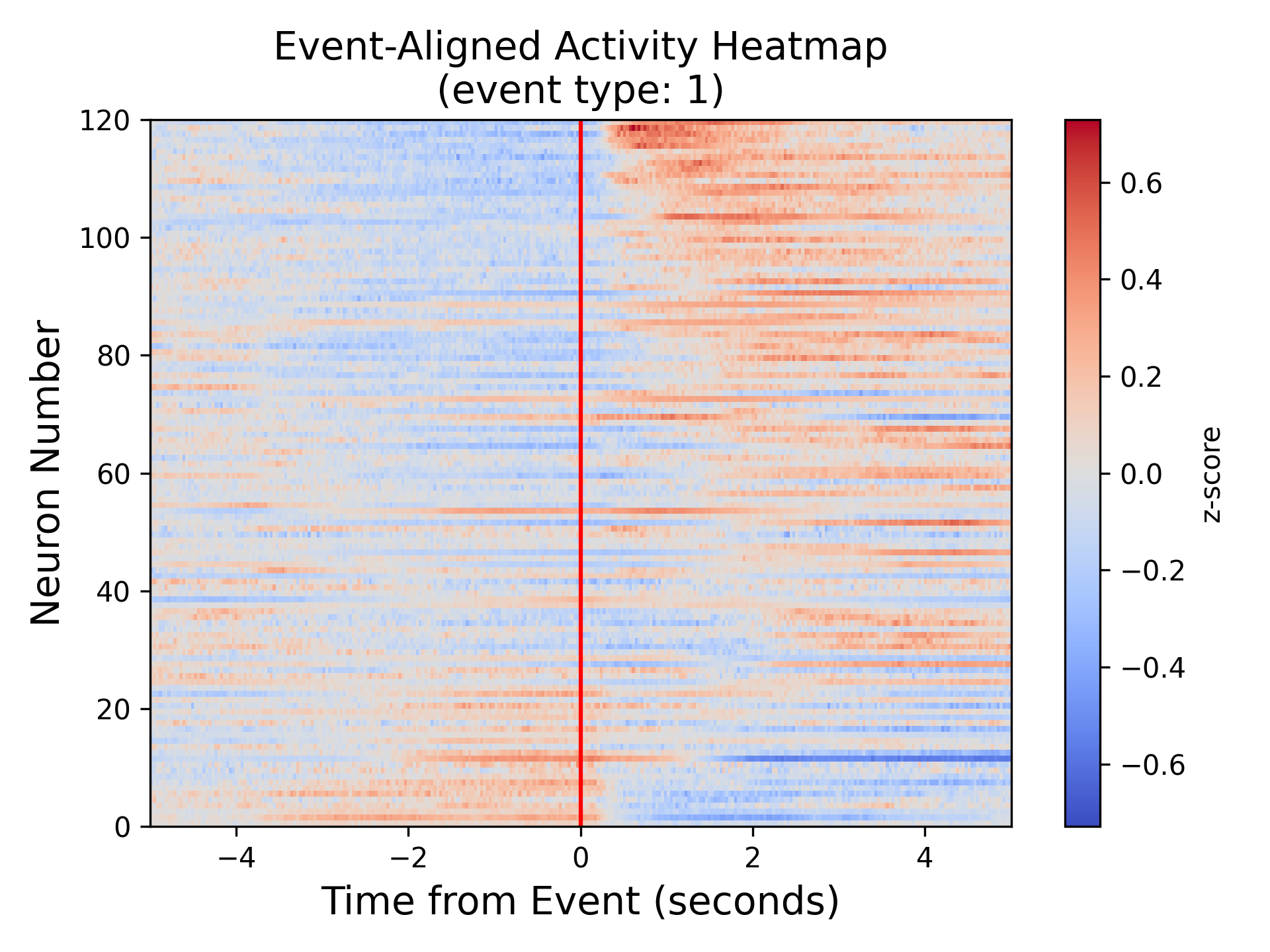
This plot shows the z-scored neural modulation around an event for each neuron in the field of view around an event (red vertical line). Warmer colors represent higher activity and cooler colors represent lower activity. The cells are sorted based on the modulation; cells are top are up-modulated whereas cells at the bottom of the plot are down-modulated. This plot captures the heterogeneity of signal which is difficult to deduce just from the analysis of pooled neural signal.
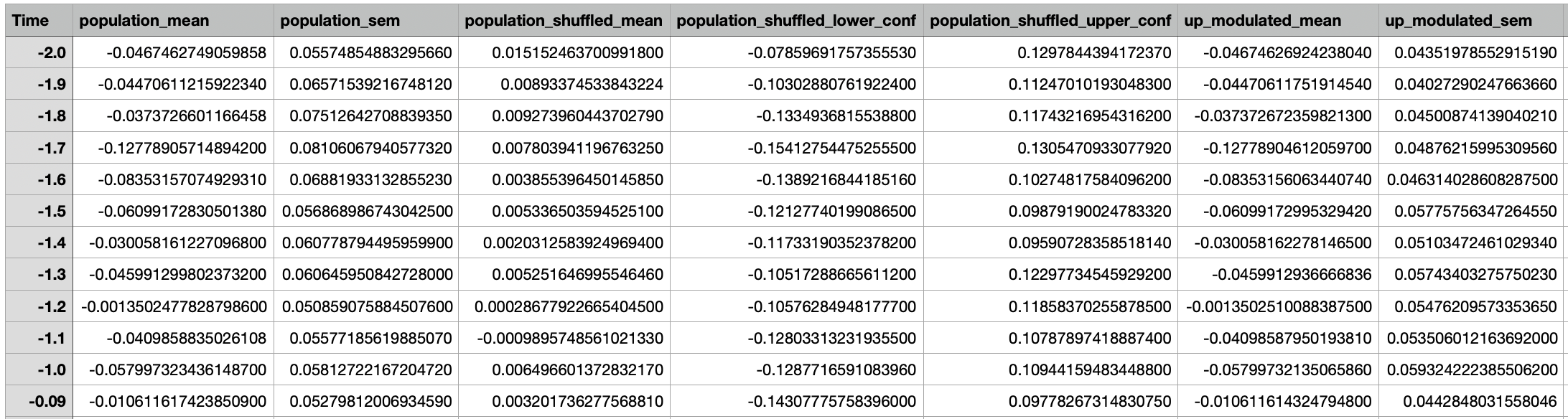
Panel B: Event-Aligned population activity¶
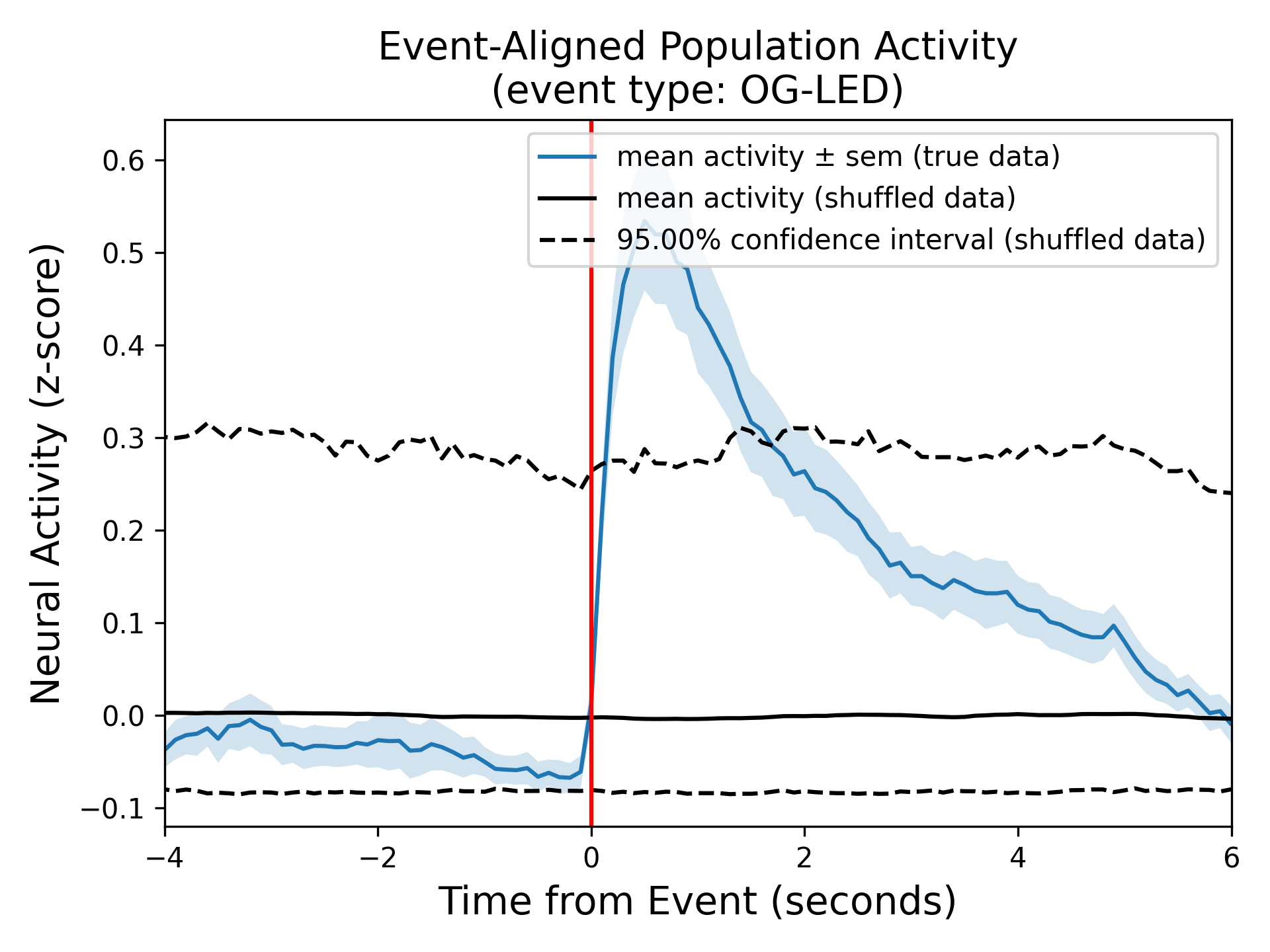
This plot provides information about the average z-scored neural activity for all neurons (blue trace) around an event (red vertical line). Specifically, the blue line represents the mean signal and the shaded light blue region represents the standard error of mean (SEM). The two black lines represent the upper and lower of the confidence interval of the null distribution obtained from the circular permutation test using shuffling, and is dictated by the ‘significance threshold’ parameter.
Panel C: Event-Aligned activity by modulation group¶
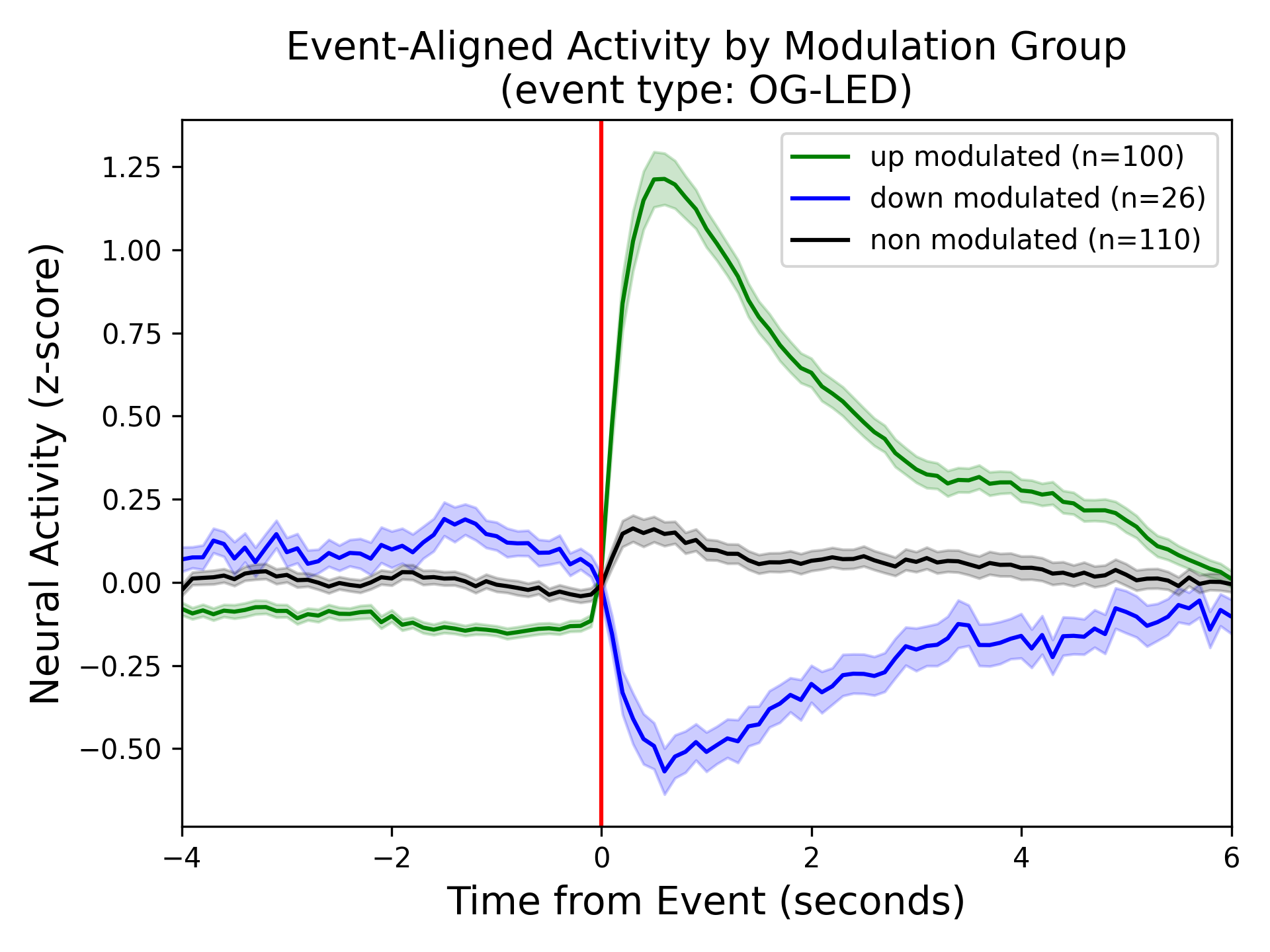
The average neural activity of the subpopulations of down-modulated (blue), up-modulated (green) and non-modulated (gray) neurons around an event (red vertical line). The line represents the mean and shaded region around the bold lines indicates the SEM.
Panel D: Cell map¶
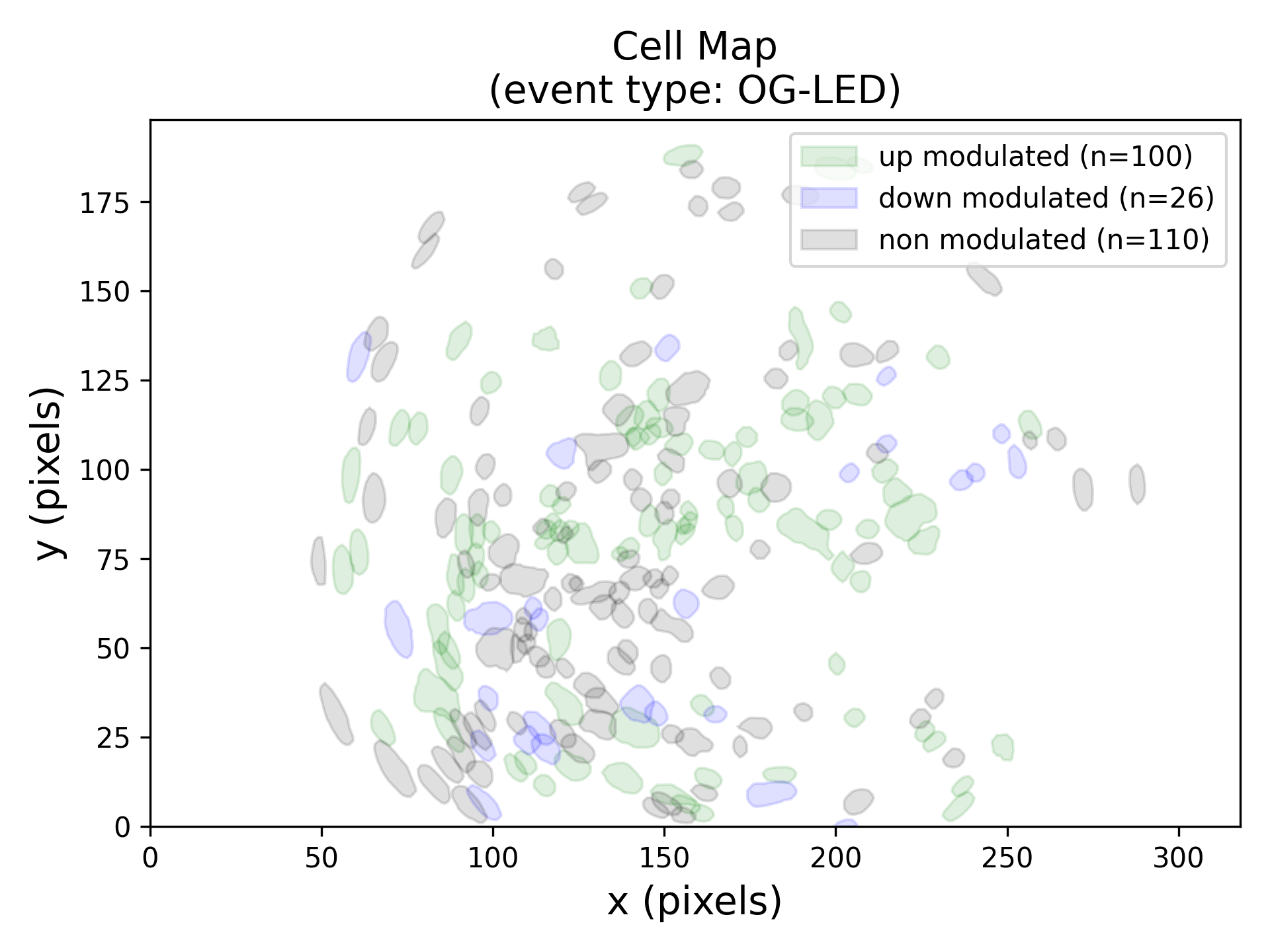
This plots provides the spatial locations of the centroids of each cell within the FOV. The color codes represent the category a cell belongs to. Blue disks are the down-modulated neurons, green disks are the up-modulated neurons and gray disks are the non-modulated neurons.
Output Data¶
For each event type, a total of 6 output files are produced. These output files can broadly fall into either one of the two categories: files containing actual data and files containing information for a preview. The (4) files containing the preview information have already been described in section Output preview The output files from the peri-event analysis workflow along with examples are described in detail below:
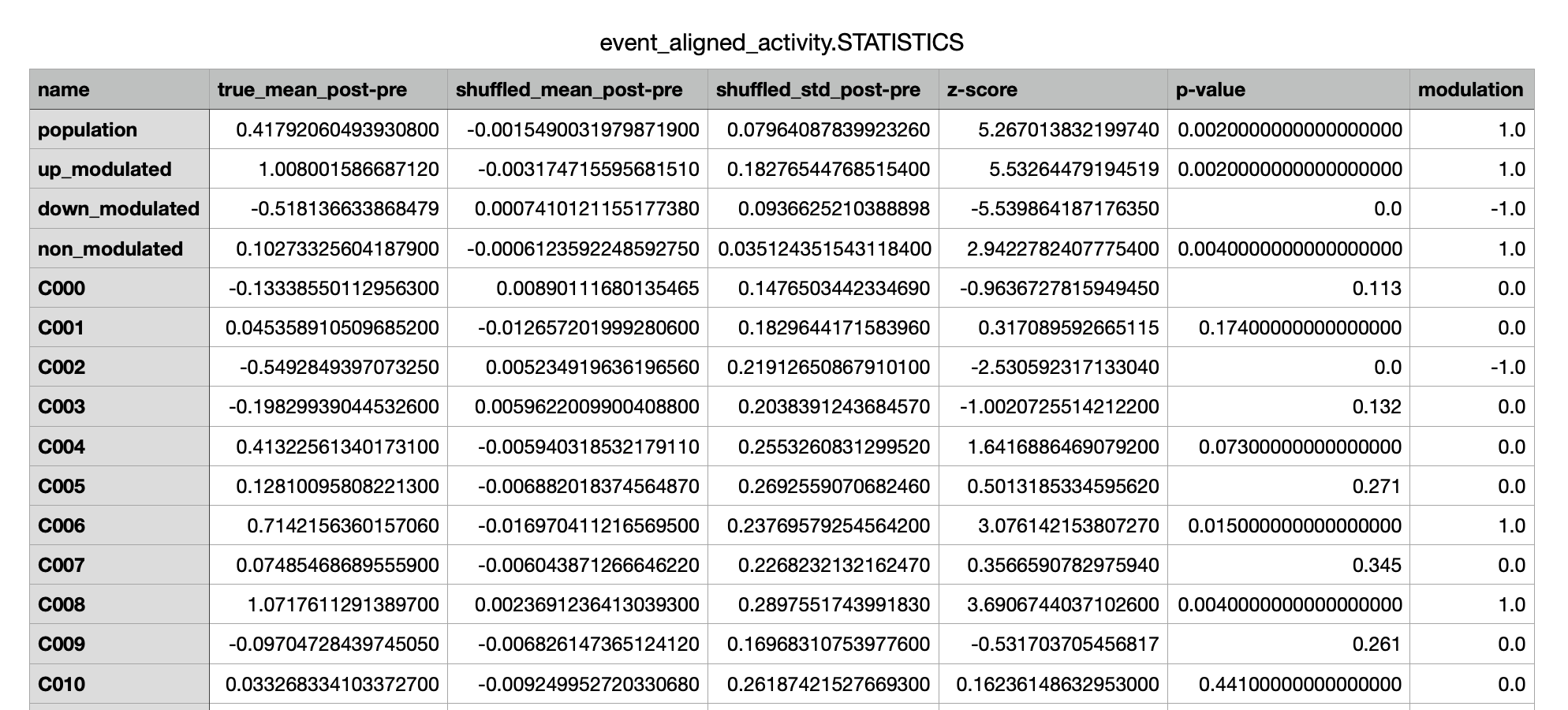
event_aligned_activity.STATISTICS.csv¶
A csv file containing the information about the output of statistical tests performed on each cell and
some its associated metrics.
event_aligned_activity.TRACES.csv¶
A csv file containing the information about the event aligned activity of all cells.
Next Steps¶
After running this tool, the outputs can be used as input to the Combine and Compare Peri-Event Analysis Data tool.
-
Baylor, DuA, B. J. Nunn, and J. L. Schnapf. "Spectral sensitivity of cones of the monkey Macaca fascicularis." The Journal of Physiology 390, no. 1 (1987): 145-160. ↩
-
Turner, Maxwell H., and Fred Rieke. "Synaptic rectification controls nonlinear spatial integration of natural visual inputs." Neuron 90, no. 6 (2016): 1257-1271. ↩
-
Krumin, Michael, Julie J. Lee, Kenneth D. Harris, and Matteo Carandini. "Decision and navigation in mouse parietal cortex." Elife 7 (2018): e42583. ↩
-
Erlich, Jeffrey C., Bingni W. Brunton, Chunyu A. Duan, Timothy D. Hanks, and Carlos D. Brody. "Distinct effects of prefrontal and parietal cortex inactivations on an accumulation of evidence task in the rat." Elife 4 (2015): e05457. ↩
-
Zolin, Aryeh, Raphael Cohn, Rich Pang, Andrew F. Siliciano, Adrienne L. Fairhall, and Vanessa Ruta. "Context-dependent representations of movement in Drosophila dopaminergic reinforcement pathways." Nature neuroscience 24, no. 11 (2021): 1555-1566. ↩
-
Roitman, Jamie D., and Michael N. Shadlen. "Response of neurons in the lateral intraparietal area during a combined visual discrimination reaction time task." Journal of neuroscience 22, no. 21 (2002): 9475-9489. ↩
-
Fink, Andrew JP, Katherine R. Croce, Z. Josh Huang, L. F. Abbott, Thomas M. Jessell, and Eiman Azim. "Presynaptic inhibition of spinal sensory feedback ensures smooth movement." Nature 509, no. 7498 (2014): 43-48. ↩
-
Mamiya, Akira, Pralaksha Gurung, and John C. Tuthill. "Neural coding of leg proprioception in Drosophila." Neuron 100, no. 3 (2018): 636-650. ↩
-
Marshall, N.J., Glaser, J.I., Trautmann, E.M. et al. Flexible neural control of motor units. Nat Neurosci 25, 1492–1504 (2022). https://doi.org/10.1038/s41593-022-01165-8 ↩