Parameter Setting Tips¶
Parameter values depend on the signal-to-noise ratio of your dataset, and the number and size of your cells. This section provides guidance and tips on how to fine-tune the CNMF-E parameters based on the results obtained by the algorithm. The default parameters may not be optimal for all datasets. As such, it is important to understand how each parameter affects processing in order to determine which ones to tweak to obtain the desired output.
Relationships between Individual Parameters and CNMF-E Processing Modules¶
The flow chart below shows how data flows through the different CNMF-E modules during processing. The advanced parameters of CNMF-E are shown in the figure below the flow chart and colored based on the module they affect. The table underneath provides additional information about the effect of each parameter on the CNMF-E algorithm.
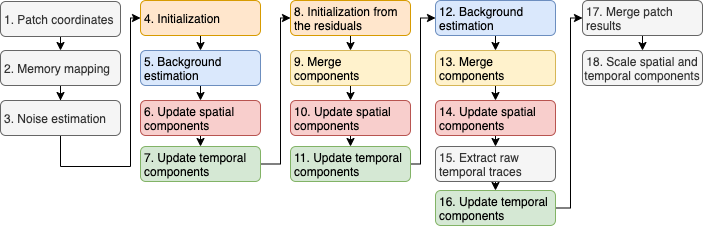
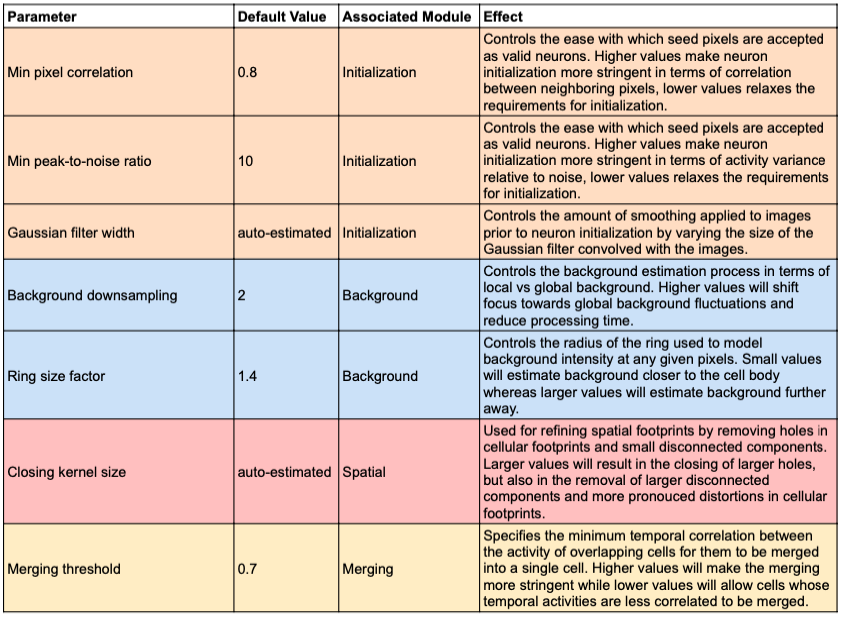
Initialization Parameters¶
Preprocessing¶
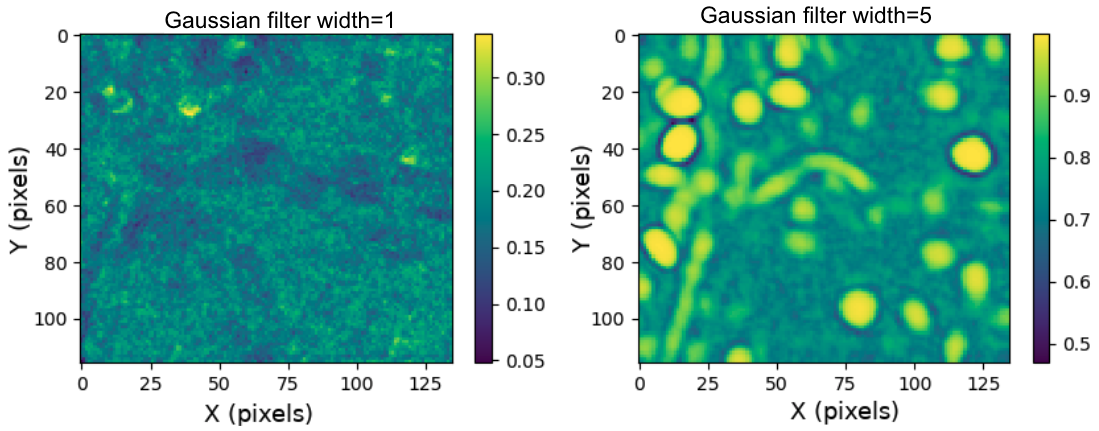
One of the first steps of the initialization module is to apply a Gaussian filter with width roughly equal to the radius of a cell in pixels. This effectively make cells pop out in the correlation image, which ultimately makes them easier to identify. An example of the effect of using different Gaussian filter sizes is shown below. In this case, a Gaussian filter width of 5 produces a crisp correlation image that leads to more identified cells. The average diameter of cells in this 4X downsampled and cropped movie is roughly 10 pixels, and a Gaussian filter width of 5 produces a crisp correlation image that leads to more identified cells.
Seed Pixels¶
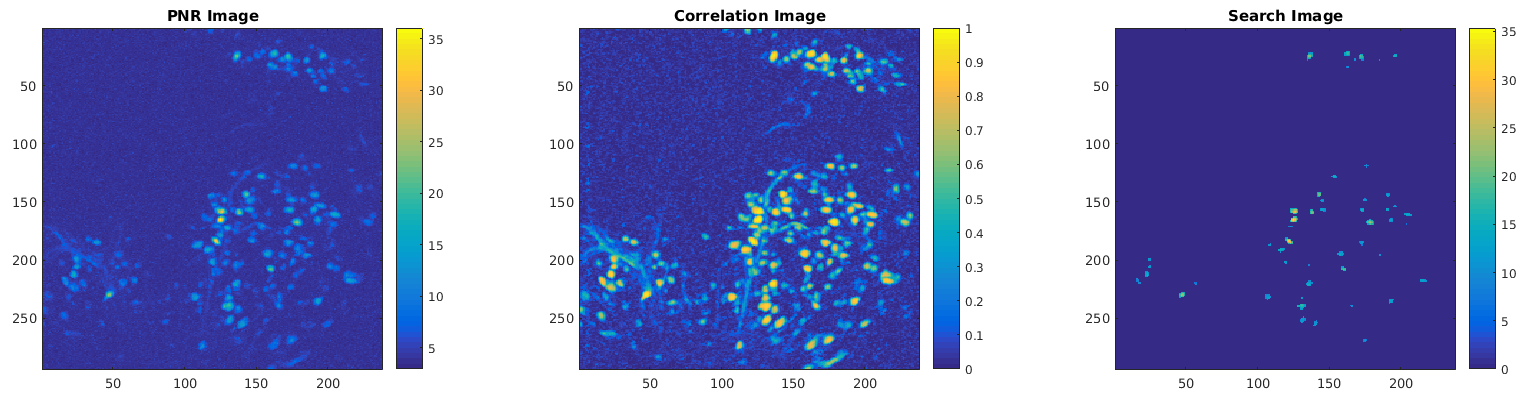
Seed pixels are pixels identified as potential neuron centers. In order for a seed pixel to actually be used to initialize the spatial footprint of a neuron, it must meet or exceed the initialization criteria. Specifically, a seed pixel must be sufficiently correlated with its immediate neighbors and must also have a level of temporal activity that exceeds some multiple of the estimated noise level. The two metrics can be computed for each pixel, providing two images that can be combined into one search image that can be used to identify neurons that meet the initialization criteria. An example of such images is displayed below: PNR Image obtained using a minimum peak-to-noise ratio of 10 (left), Correlation Image obtained using a minimum correlation of 0.8 with 8 immediate neighbor pixels (middle), and a Search Image obtained by multiplying the PNR Image with the Correlation Image (right).
Background Parameters¶
Downsampling¶
Spatial downsampling is used when estimating the background to reduce the effects of local fluctuations on the background estimation. The amount of downsampling thus control the balance between local and global background. Larger downsampling values will result in a background estimation that is primarily dominated by global fluctuations whereas lower downsampling values will place more emphasis small variations in the background across the field of view. Since the background estimation procedure is one of the most expensive operations in the CNMF-E algorithm, larger downsampling values can significantly improve processing speed.
Background Model¶
The background fluorescence is estimated for every pixel using a ring model, i.e. by considering the intensity of pixels that form a ring around any given pixel. The radius of the ring controls how many pixels to use to estimate the background and how far from the current pixel to look for background fluctuations.
Spatial Parameters¶
The spatial module is responsible for processing and refining the spatial footprints of the neurons identified by the algorithm. The closing kernel size is an input parameter that controls the morphological closing operation applied to each footprint during processing. It enables the closing of small holes that may be visible in the center of a cellular footprint that has a doughnut shape. The closing operation can also be used to remove small disconnected components from cellular footprints.
Merging Parameters¶
One advantage of CNMF-E is that the cell identification process allows for overlapping cells to be identified. However, in some cases a single cell may be identified as two overlapping regions during processing. The merging module is responsible for merging such regions together into whole cells based on the correlation between their temporal activities. More specifically, the merging threshold parameter specifies the minimum temporal correlation required for two overlapping components to be merged into a single one. If the algorithm appears to oversegment cells, then one could benefit from relaxing the merging criteria by reducing the merging threshold.
Processing Parameters¶
Processing Modes¶
Three processing modes are offered for CNMF-E, each with their own memory-speed tradeoff. When CNMF-E runs in patch mode, it breaks the movie into overlapping patches and does cell identification on the individual patches, and merges the results in the end. Under this mode, the processing time and the amount of memory consumed by CNMF-E can be controlled in part by specifying the patch size, patch overlap, and number of threads.
The parallel patch mode offers faster processing at the cost of a larger memory footprint. Its speed advantage comes from processing smaller portions of the field of view in parallel. Under this mode, the Number of threads parameter dictates how many patches can be processed in parallel at any given time.
The sequential patch mode consumes the least amount of memory, but can take significantly longer to complete processing. Under this mode, the Number of threads parameter controls how many threads will be used for operations that can be done in parallel while processing patches one at a time.
All-in-memory mode is also slower than parallel patch mode and requires a large amount of memory. The different processing modes are depicted in the figure below.
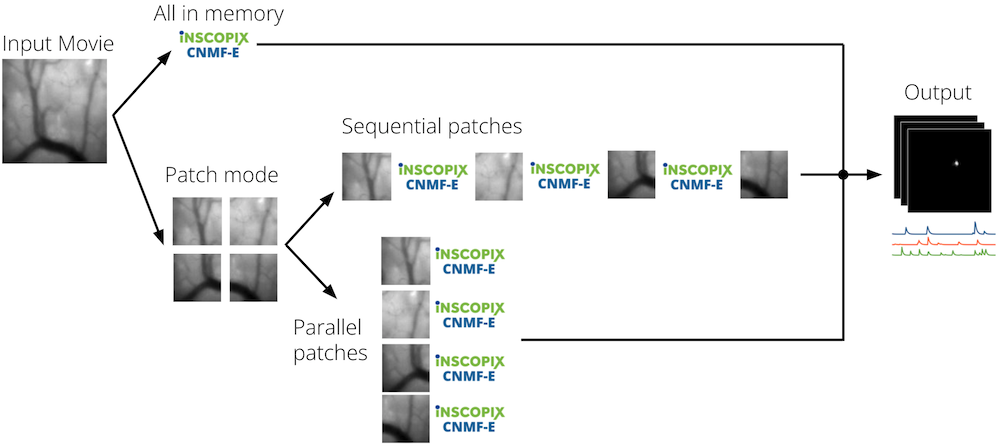
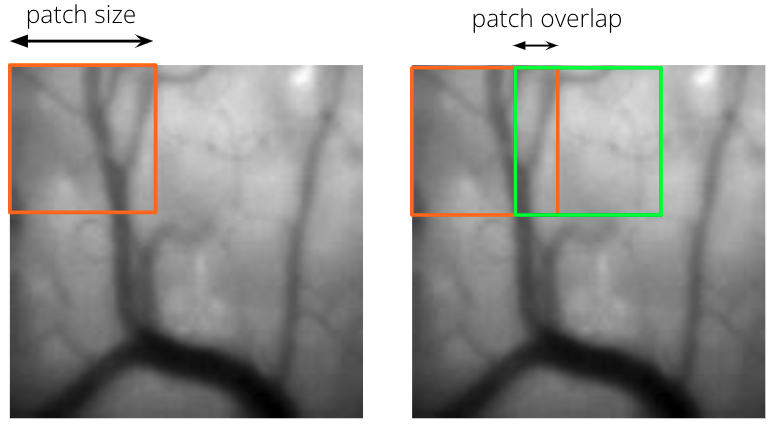
Patch Size and Patch Overlap¶
Patch size refers to the side length of an individual square patch of the field of view in pixels. Patch overlap refers to the amount of overlap between adjacent patches in pixels. These two concepts are illustrated below.
Number of Threads¶
The number of threads controls the amount of parallelism used during processing. When using all-in-memory or sequential patch mode, increasing the number of threads will result in a higher level of parallelism in the individual CNMF-E processing modules. However, a larger number of threads will not always lead to faster processing. Specifically, if the number of threads exceeds the number of cores available on your machine you may start seeing a plateau or even an increase in processing time as the overhead of managing the extra threads start exceeding the benefits of parallelization. When using parallel patch mode, the number of threads will instead control the number of patches being processed in parallel at any given time. In this mode, each thread will process one patch at a time, each time grabbing the next patch to process in the queue until all patches are processed.
Auto-Estimated Parameters¶
Inscopix CNMF-E is able to automatically estimate an appropriate value for some of the input parameters based on the specified average cell diameter. Specifically, the Gaussian filter size and closing kernel size can both be automatically estimated using the following formula, where \(f\) refers to the filter size and \(d\) the average cell diameter:
Parameter explorer¶
The following widget allows you to explore the effect of changing some parameters on an example dataset.