Common Issues¶
Common issues with the CNMF-E algorithm are described in this section along with guidelines for addressing them.
Inaccurate Average Cell Diameter¶
This parameter should not be deliberately under- or over-estimated as this could affect the quality of the output. The user should instead try to adjust the other parameters based on the tips provided in this section.
Missing Cells¶
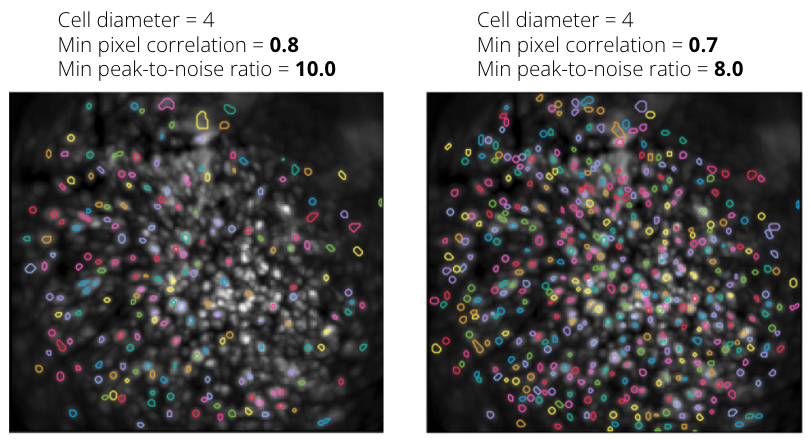
If you feel that CNMF-E missed some cells that are visible in the field of view, try relaxing the initialization parameters while maintaining the other parameters constant. More specifically, try reducing the minimum pixel correlation and peak-to-noise ratio. Note that it is not recommended to specify an inaccurate average cell diameter in an attempt to capture the missing cells as this could impair the quality of the output obtained from CNMF-E. Below is an example showing the effect of relaxing the initialization criteria. Some cells visible in the field of view were missed when using a minimum pixel correlation of 0.8 and a minimum peak-to-noise ratio of 10 (left), but were identified when relaxing the initialization criteria (right).
Too Many Cells¶
If you feel that CNMF-E identified too many cells, specifically blobs that don't appear to be cells, try making the initialization parameters more stringent by increasing the minimum pixel correlation and the peak-to-noise ratio. Also, make sure to use the appropriate average cell diameter as underestimating or overestimating this value could lead to the identification of additional blobs that may not be cells of interest.
Oversegmented Cells¶
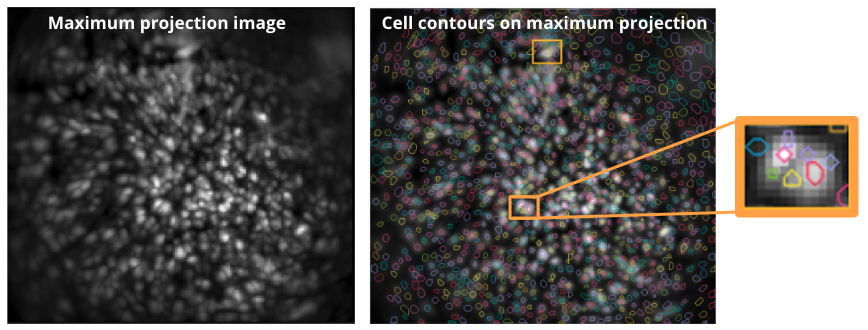
If you find that CNMF-E is overfragmenting cells, i.e. the same cell appears to be identified as multiple distinct components, try reducing the merging threshold to allow CNMF-E to combine such fragments together during processing. Again, make sure to use an appropriate average cell diameter as underestimating it could lead to the identification of smaller blobs within cells. Below is an example showing cells that were oversegmented and identified as distinct cells. In this case it is best to validate the average cell diameter and reduce the merging threshold to prevent such oversegmentation of the cell bodies.
Slow Processing¶
Processing time is largely affected by the input dimensions, the chosen processing mode, and the parallelism associated with the specified processing parameters. Parallel patch mode is the fastest processing mode, but it requires more memory. If you notice while running CNMF-E that you still have a large amount of unused memory available on your machine, try increasing the number of threads used with parallel patch mode. Background estimation is one of the most expensive operations in CNMF-E. As such, increasing the background downsampling factor can also significantly reduce processing time while shifting the background estimate toward a global rather than a localized estimate. Another way to improve processing speed is to downsample the input data as per our recommended workflow. Many operations are performed on every pixel. Downsampling can help reduce noise and also dramatically reduce processing time.