DeepLabCut Pose Estimation¶
This tool uses 2.0 compute credits per hour.
DeepLabCut is an open-source library for pose estimation based on deep learning. This tool allows users to upload any pre-trained DeepLabCut model and perform pose estimation on behavioral movies. The code used to run this tool can be found here.
Parameters¶
| Parameter | Required? | Default | Description |
|---|---|---|---|
| Zipped Model Folder | True | N/A | DeepLabCut model folder compressed in a .ZIP file. The contents of this file will be extracted and used for execution with DeepLabCut |
| Behavior Movie | True | N/A | Behavioural movies to analyze. Must be one of the following formats: .isxb, .mp4, and .avi. |
| Experiment Annotations Format | True | parquet | The file format of the output experiment annotations file. Can be either .parquet or .csv |
| Crop Rectangle | False | N/A | Draw a cropping rectangle on the input movie which will be used by DeepLabCut to crop movie frames before running the model. |
| Window Length | True | 5 | Length of the median filter applied on the predictions. Must be an odd number. If zero, then no filtering is applied. |
| Displayed Body Parts | True | all | Selects the body parts that are plotted in the video. The body parts are specified in the config.yaml file of the zipped model folder. |
| P Cutoff | True | 0.6 | Cutoff threshold for predictions when labelling the input movie. If predictions are below the threshold, then they are not displayed. |
| Dot Size | True | 5 | Size in pixels to draw a point labelling a body part. |
| Color Map | True | rainbow | Color map used to color body part labels. Any matplotlib colormap name is acceptable. |
| Keypoints Only | True | False | Only display keypoints, not video frames. |
| Output Frame Rate | False | N/A | Positive number, output frame rate for labeled video. If None, use the input movie frame rate. |
| Draw Skeleton | True | False | If True adds a line connecting the body parts, making a skeleton on each frame. The body parts to be connected and the color of these connecting lines are specified by the Color Map. |
| Trail Points | True | 0 | Number of previous frames whose body parts are plotted in a frame (for displaying history). |
Inputs¶
The following table summarizes the valid input files types for this tool:
| Source Parameter | File Type |
|---|---|
| Zipped Model Folder | Any [.zip] |
| Behavior Movie | nVision Movie [.isxb] or Movie (general) [.mp4, .avi] |
The following sections explain in further detail the expected format for these input files.
Zipped Model Folder¶
A DeepLabCut model is stored as a folder with specific content and structure.
This folder is expected to compressed in a .zip file.
At the start of tool execution, the contents of zipped model folder are examined in order to detect whether it can be used with DeepLabCut.
Direct Usage with DeepLabCut¶
First, the tool checks whether the contents of the zipped model folder can be used directly with DeepLabCut. This check examines if the contents of the zipped model folder has a directory structure which matches the directory structure of DeepLabCut projects. A DeepLabCut project is a folder created by DeepLabCut when models are trained. Subsequently, the same folder can be used by DeepLabCut to analyze new behavioral videos. A DeepLabCut project has the following directory structure:
config.yamldlc-modelsiteration-<iteration><task>-<date>-trainset<trainingfraction>shuffle1testpose_cfg.yaml
trainpose_cfg.yamlsnapshot-<snapshotnumber>.data-00000-of-00001snapshot-<snapshotnumber>.indexsnapshot-<snapshotnumber>.meta
The config.yaml file is generated by DeepLabCut, storing the configuration of the DeepLabCut project that was used to train models. This configuration includes the list of body parts in the animal pose, the skeleton of the animal pose, and the type of neural network used for training.
The dlc-models folder contains the DeepLabCut models trained in the project.
Iterations of model training are stored in sub-folders.
The value <iteration> is read from config.yaml, which determines the iteration to use for analysis.
By default this is set to 0 in config.yaml.
Inside the folder iteration-<iteration>, there should be a sub-folder named <task>-<date>-trainset<trainingfraction>shuffle1.
The values <task>, <date>, and <trainingfraction> are all read from config.yaml as well.
Within the folder <task>-<date>-trainset<trainingfraction>shuffle1, there should be two sub-folders named train and test.
The train folder contains files used & generated during model training.
The test folder contains files used & generated during model testing.
Both folders should contain a configuration file named pose_cfg.yaml.
The train folder should also contain snapshot files of the model captured during training iterations.
A snapshot generally consists of three files .data-00000-of-00001, .index, .meta.
Ideally, there should only be files for one snapshot number, indicated by <snapshotnumber>, since only one snapshot of the model is actually used for analysis.
The snapshot number to use is indicated by snapshotindex in config.yaml (index of unique snapshot numbers in the training directory).
If more than one unique snapshot number is found in the train folder, a warning is logged by the tool.
Save on space
Ensure there's only one snapshot in the zipped model folder before uploading to IDEAS. Snapshot files make up the majority of the size of a DeepLabCut model. Including only one snapshot ensures the size of the zipped model folder uploaded to IDEAS is as small as possible, saving on storage space.
Indirect Usage with DeepLabCut¶
If the contents of the zipped model folder do not correspond to the structure of a DeepLabCut project, the tool will try to construct a new DeepLabCut project and copy the relevant files in the zipped model folder to the new project.
The tool recursively searches for a sub-folder that contains snapshot files (i.e., file name starts with snapshot).
Snapshot ambiguity
If there's more than one folder with snapshot files the tool will fail because it's unable to determine which folder to use for analysis.
If more than one unique snapshot number is found in the snapshot folder, a warning is logged.
Save on space
Ensure there's only one snapshot in the zipped model folder before uploading to IDEAS. Snapshot files make up the majority of the size of a DeepLabCut model. Including only one snapshot ensures the size of the zipped model folder uploaded to IDEAS is as small as possible, saving on storage space.
Otherwise, in the snapshot folder, the tools checks if there is a pose_cfg.yaml file.
The tool then creates a new project and copies the following values from pose_cfg.yaml in the zipped model folder to config.yaml in the new project:
default_net_type -> net_typedataset_type -> default_augmenterall_joints_names -> bodyparts
Moreover, the tool copies the following keys from pose_cfg.yaml in the zipped model folder to the pose_cfg.yaml files in the test and train folders of the new project:
datasetdataset_typenum_jointsall_jointsall_joints_namesnet_typeinit_weightsglobal_scalelocation_refinementlocref_stdev
This mapping of keys from pose_cfg.yaml to the config files in the new DeepLabCut project is based on this logic in the DeepLabCut Model Zoo API.
Behavior Movie¶
The tool will analyze behavioral movies using the zipped model folder.
The behavior movies can be in the one the following file formats: .isxb, .mp4, .avi.
Multiple input movies
The tool can accept multiple behavior movies (of the same file format) as input, applying the model on each movie individually.
If the behavior movies are in .isxb file format, then the movies can be constructed into a series on IDEAS and then used as input to the tool as well.
See the Outputs section for more information on how outputs are formatted for multiple input movies.
Algorithm Description¶
Once the tool has validated that it can use the zipped model folder input with DeepLabCut, the analysis begins. This tool consists of three main analysis steps, summarized in the following diagram:
The first step is running the deeplabcut.analyze_videos API function, documented here.
This function takes a path to the DeepLabCut project which contains the trained model to use, and applies that model on the input behavior movies.
The output of this step is an h5 file containing the pose estimates of the model.
The second step is running the deeplabcut.filter_predictions API function, documented here.
This functions filters the raw pose estimates, removing outliers and noise from the results.
The output of this step is also an h5 file containing the filtered pose estimates of the model.
This step is recommended in the DeepLabCut documentation, however it can be skipped by setting the Window Length parameter to one.
The third and final step is running the deeplabcut.create_labeled_videos API function, documented here.
This function creates a video of the input movie with annotations of the pose estimates results.
If filtering is applied on the pose estimates, it will annotate the input movie with the filtered results.
Otherwise, the raw pose estimates will be used for annotations.
The output of this step is a mp4 file with the annotated movie frames.
This result helps to easily visualize and assess the quality of the model results.
Outputs¶
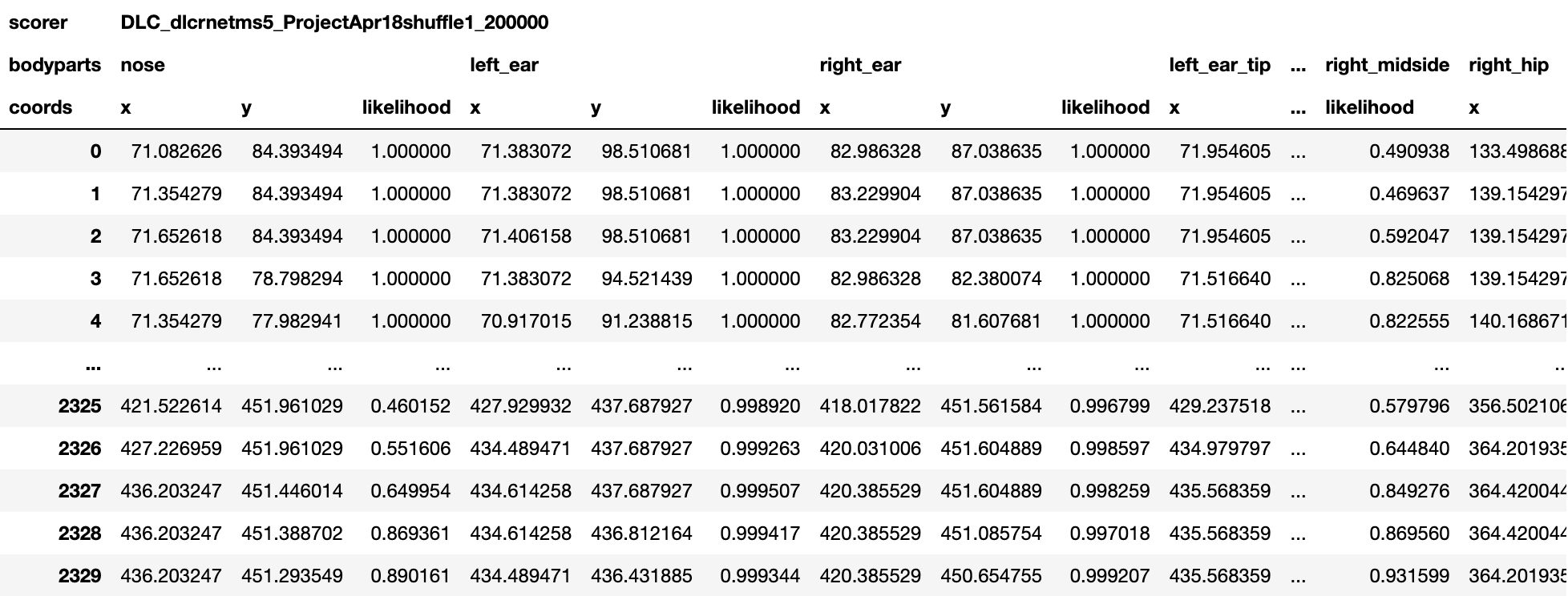
Pose Estimates H5 File¶
An .h5 file containing the model predictions.
The predictions are stored as a MultiIndex Pandas Array.
The files contains the name of the network, body part name, (x, y) label position in pixels, and the likelihood (i.e., how confident the model was to place the body part label at those coordinates) for each frame per body part.
The following figure shows an example of this output.
Filtering
If the Window Length parameter is set to one, then no filtering is performed and the raw pose estimates (aka coordinates or keypoints) are output.
Otherwise if the Window Length is greater than one, then filtering is performed and the filtered pose estimates are output.
Multiple input movies
If there are multiple input movies, a pose estimates.h5 file is created for each movie.
Previews¶
Each pose estimates file will have the following previews to visualize the data stored in these files.
All of these figures are generated using the plot_trajectories DeepLabCut API function.
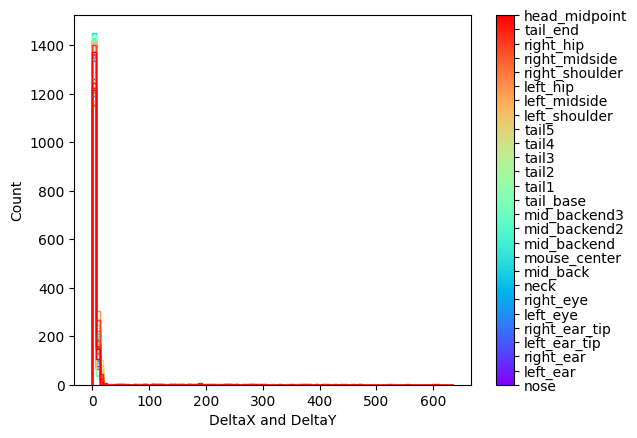
Histogram:
The first preview is a histogram plot of the consecutive differences in x and y coordinates for each body part. This plot can help determine if there's any outliers in the results based on significant jumps in the coordinates of a body part. Ideally the distribution for each body part should be close to zero.
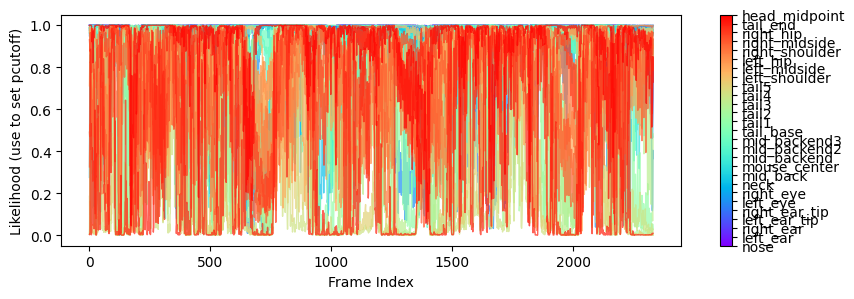
Likelihood vs. Time Plot:
The second preview is plot of the likelihood of each body part over time. This plot can help identify body parts or time periods where likelihood is low, indicating less reliable results from the model to filter in subsequent analysis.
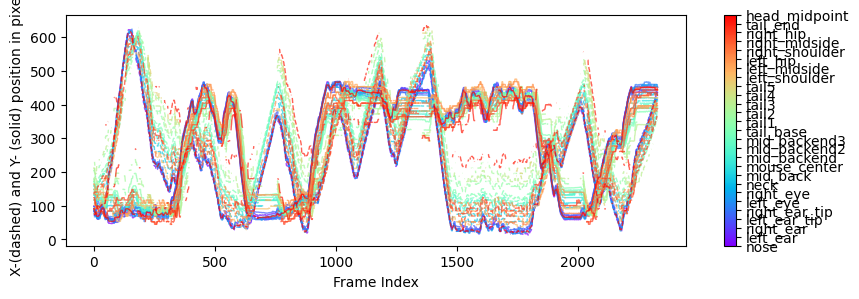
Body Parts vs. Time Plot:
The third preview is a plot of the x & y coordinates of each body part over time. This plot can help identify how the position of body parts changes over time.
Body Parts Trajectory:
The fourth preview is a plot of the x & y coordinates of each body part plotted spatially in the FOV (field of view). This plot can help identify where the location of body parts in the FOV throughout the duration of the input movies.
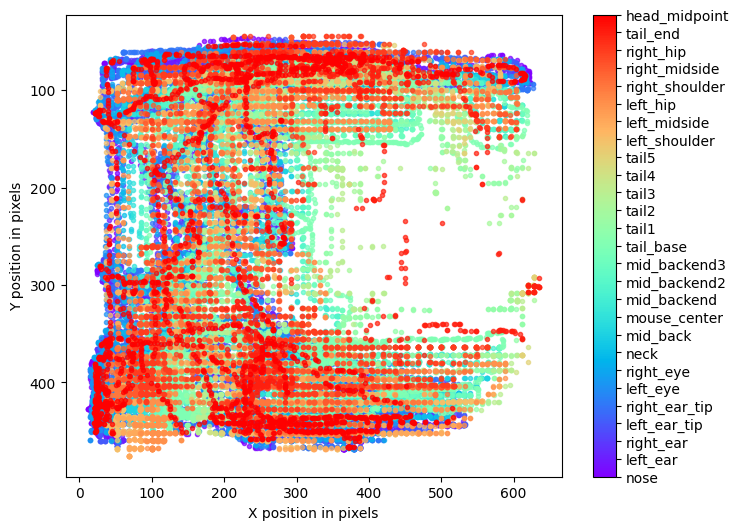
Experiment Annotations File¶
The pose estimates in IDEAS experiment annotations format. This can be either a .csv or .parquet file depending on the Experiment Annotations Format parameter. The following table shows an example of this output.
| Frame number | Movie number | Local frame number | Time since start (s) | Hardware counter (us) | nose x | nose y | nose likelihood | ... | head_midpoint x | head_midpoint y | head_midpoint likelihood |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | 0 | 0.0 | 90310284262 | 71.08261108398438 | 84.39350891113281 | 0.9999998807907104 | ... | 71.38306427001953 | 98.51067352294922 | 0.9999996423721313 |
| 1 | 0 | 1 | 0.033333 | 90310352280 | 71.3542709350586 | 84.39350891113281 | 0.9999998807907104 | ... | 71.38306427001953 | 98.51067352294922 | 1.0 |
| 2 | 0 | 2 | 0.066666 | 90310420240 | 71.6526107788086 | 84.39350891113281 | 0.9999998807907104 | ... | 71.40615844726562 | 98.51067352294922 | 1.0 |
| 3 | 0 | 3 | 0.099999 | 90310484246 | 71.6526107788086 | 78.79830932617188 | 0.9999998807907104 | ... | 71.38306427001953 | 94.52143859863281 | 1.0 |
| 4 | 0 | 4 | 0.133332 | 90310552256 | 71.3542709350586 | 77.9829330444336 | 0.9999995231628418 | ... | 70.91700744628906 | 91.23883056640625 | 1.0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
For every body part estimated by the model, there are three columns that are output: x, y, and likelihood. In addition, the following columns are included:
Frame number: The frame number in the input movie. If there are multiple input movies, this column will refer to the global frame number across the frames in all the input movies.Movie number: The movie number that the frame is from, within the multiple input movies.Local frame number: The frame number within the individual movie that the frame is from.Time since start (s): Timestamp in seconds for every frame of the input movie, relative to the start of the movie.Hardware counter (us): Only included for.isxbinput movies. This column contains the hardware counter timestamps in microseconds for every frame of the input movie. These are the hardware counter values generated by the nVision system which can be compared to corresponding hardware counter values in.isxd,.gpio, and.imufiles from the same synchronized recording. These timestamps will be used downstream in theMap Annotations to ISXD Datatool in order map frames from annotations to isxd data, so the two datasets can be compared for analysis.
Multiple input movies
If there are multiple input movies, there will still only be one experiment annotations file, which concatenates the pose estimates across all input movies into a single result.
Labeled MP4 File (for visualization)¶
A .mp4 file containing the model predictions annotated on every frame of the input movie.
The following figure shows an example of this output.

Multiple input movies
If there are multiple input movies, a labeled movie .mp4 file is created for each movie.
Next Steps¶
Here are some examples of subsequent analyses that can be executed using the outputs of this tool:
- Average DeepLabCut Keypoints: Average the keypoint estimates in each frame of the tool output, and use this to represent the mouse center of mass (COM).
- Compute Locomotion Metrics: Compute instantaneous speed and label states of rest and movement from averaged keypoints.
- Compare Neural Activity Across States: Compute population activity during states of rest and movement from locomotion metrics.
- Compare Neural Circuit Correlations Across States: Compute correlations of cell activity during states of rest and movement from locomotion metrics.
- Combine and Compare Population Activity Data: Combine and compare population activity from multiple recordings and compare across states of rest and movement.
- Combine and Compare Correlation Data: Combine and compare correlations data from multiple recordings and compare across states of rest and movement.